ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2015) Volume 26, Issue 4
White Blood Cells Classifications by SURF Image Matching, PCA and Dendrogram
1Department of Mechatronics Engineering, Faculty of Engineering, Atilim University, 06836, Ankara, Turkey
2Department of Electronics Engineering Department, Faculty of Engineering, Ankara University, 06100, Ankara, Turkey
3Department of Information Systems Engineering, Faculty of Engineering, Atilim University, 06836, Ankara, Turkey
4Department of Computer Engineering, Faculty of Engineering, Atilim University, 06836, Ankara, Turkey
5Department of Computer Engineering, Faculty of Engineering, Yasar University, 35100, Izmir, Turkey
- *Corresponding Author:
- Gökhan Şengül
Computer Engineering
Faculty of Engineering
Atilim University, 06836
Ankara, Turkey
Accepted date: July 2015
Determination and classification of white blood cells are very important for diagnosing many diseases. The number of white blood cells and morphological changes or blasts of them provide valuable information for the positive results of the diseases such as Acute Lymphocytic Leucomia (ALL). Recognition and classification of white cells as basophils, lymphocytes, neutrophils, monocytes and eosinophils also give additional information for the diagnosis of many diseases. We are developing an automatic process for counting, size determination and classification of white blood cells. In this paper, we give the results of the classification process for which we experienced a study with hundreds of images of white blood cells. This process will help to diagnose especially ALL disease in a fast and automatic way. Three methods are used for classification of five types of white blood cells. The first one is a new algorithm utilizing image matching for classification that is called the Speed-Up Robust Feature detector (SURF). The second one is the PCA that gives the advantage of dimension reduction. The third is the classification tree called dendrogram following the PCA. Satisfactory results are obtained by two techniques.
Keywords
ALL disease, white blood cell, SURF, PCA, NN, Dendrogram
Introduction
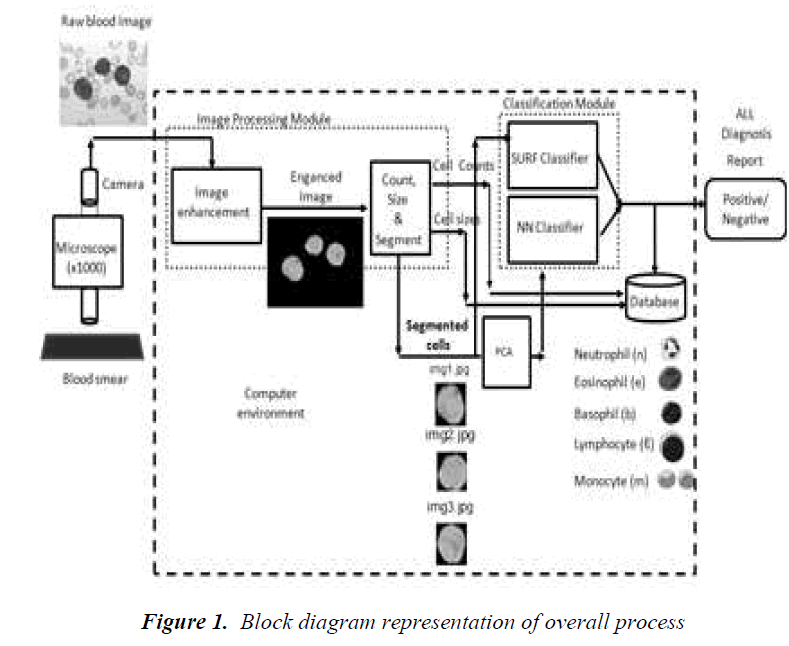
The motivation behind this study is to develop an automatical process for helping the diagnosis of Acute Lymphocytic Leucomia (ALL) desease. Most of the diseases can be diagnosed by the numbers and sizes of white blood cells found in a blood smear. Among other diseases, ALL disease that has to be diagnosed in a fast and accurate way is very critical for the health of children. Today, manual methods are used to count and cell size determination. This can give rise to inaccurate results. It is also very tedious effort to determine number of cells within a blood smear. The results are dependent on the situation of the expert doing the analysis. Therefore, a reliable automatic and fast way is vital for determining number of cells and sizes of them. The main purpose of this paper is to describe the development and to present the results of a blood smear image based process to help for diagnosis of diseases. Recognition and classification of the types of white blood cells as basophils, lymphocytes, neutrophils, monocytes and eosinophils also give additional information for the diagnosis of many diseases. We are developing an automatical process for counting, size determination and classification of white blood cells. In this paper, we give mainly the results of the developed classification process. It is the core of a computer aided diagnosis system. This process will help to diagnose especially ALL disease in a fast and automatic way. The process may replace the classical manual process that is still used in medical laboratories today. Since the process that we developed is a computer based system, it can give us invaluable opportunities for diagnosis and treatment of diseases. The process is mainly an image based system. There is no need for using blood itself during the process. The experiments and analysis can be repeated easily and frequently. The results can be obtained in a fast way and they are reliable and accurate enough. A database can be created for patients. It can be reached by the doctors when they need to see the state of a patient in any time. The system can be the part of a computer network in a hospital. Remote access to the system and its database can be possible. Diagnosis and treatment can be achieved quickly. The process that we developed has two main sub-processes (Fig.1). The first one is the image processing part and the second is the classification part. Image processing is necessary for segmenting individual white cells for further processing such as determination of the number of cells and sizes of cells, and classification as well. Three methods are used for classification. The first one is a new algorithm utilizing image matching for classification that is called the Speed-Up Robust Feature detector (SURF). The second one is to use PCA for dimension reduction purpose, a couple of classical artificial neural network structures. We applied principal component analysis for classification. A classification tree is also created following the PCA application. It is called the dendrogram obtained for the types of white blood cells.
Diseases can be diagnosed by the number and morphological changes of white blood cells. Today, the diagnosis has still been achieved mainly by manual techniques. However, the accuracy of it depends on the operator’s expertise. The situation of the operator may highly affect the analysis. Recently, there are efforts and research studies on making it automatic the process [1-3]. The works on automatization can be divided into two parts, namely, segmentation and classification of blood cells. In this paper, we mainly concentrated on the classification problem. Since there are a lot of cells in a blood smear, we have to find a suitable way to detect and classify them. In literature, there are some effective methods for classifying high dimensional data [4,5]. We apply principal component analysis (PCA) for reducing high dimensional data. Neural networks are also widely used for classification of white blood cells [6,7]. Segmentation of cells is also one of the main topics on the white blood cell analysis. Shape is an important characteristic for determining a lot of diseases including ALL. Therefore, both red and white cell shape estimation are studied [8,9] K-mean clustering method and Fuzzy Cmean clustering method are widely used in segmenting white blood cells[10,11]. The studies in [12,13], the authors developed robust segmentation and measurement techniques of white cells in blood microscope images. They tried to identify ALL diseases. In the literature, there are a couple of studies on blood microscopic image segmentation and automated identification and classification of white blood cells utilizing different methods [14-16]. Automatic and semiautomatic white blood cell segmentation studies are given in [17,18].
Subjects and Methods
In our work, a new and completely automatic segmentation and classification process is developed. In this paper, the contribution of our work is the introduction of a new algorithm for image classification that is called the Speed-Up Robust Feature detector (SURF) for the classification problem of white blood cells. This algorithm is effective for scale invariant feature transform. Our approach does not need to extract nucleus and cytoplasm. We utilize image matching in this method. We make use of image matching as the classification purpose. We also use the original image after PCA application and training of neural networks. Hierarchical clustering that is represented by a tree called dendrogram is used for classification validation the cells. In our case we don't need any expertise because of automatic thresholding during segmentation by Otsu's method
Pre-Processing
The overall process is given in Figure 1. It consists of some important stages. The target process is aimed to produce the following outputs: (1) the number of white blood cells within the image; (2) the sizes of individual white blood cells; (3) the percentage of malignant (grown) white blood cells called lymphoblasts; (4) the classes of the white blood cells; and (6) the diagnosis of Acute Lymphocytic Leukemia (ALL) disease giving positive or negative answer (this part is out of the scope of this study). The image processing method applied here has some drawbacks. One of them is to be able to extract completely occluded cells and the other is to distinguish
cells which stick together. These cells are much greater than the others since the connected components labeled during the process may actually have two or more cells rather than one cell. In such a situation, the count number will be erroneous. However, since we check the ratios of both axes of the cells, we can easily realize that the cells that have ratios greater than 100% are partly occluded by the others or they are so close that they touch together. In that case, although the algorithm counts them as a single cell, we correct the count number by increasing the counter by one if the ratio is in between 100% and 200%. We increment the counter by two if the ratio is greater than 200%. Normally this is enough in most of the applications. No manual intervention was needed for the experiments carried out in the above applications.
Classification
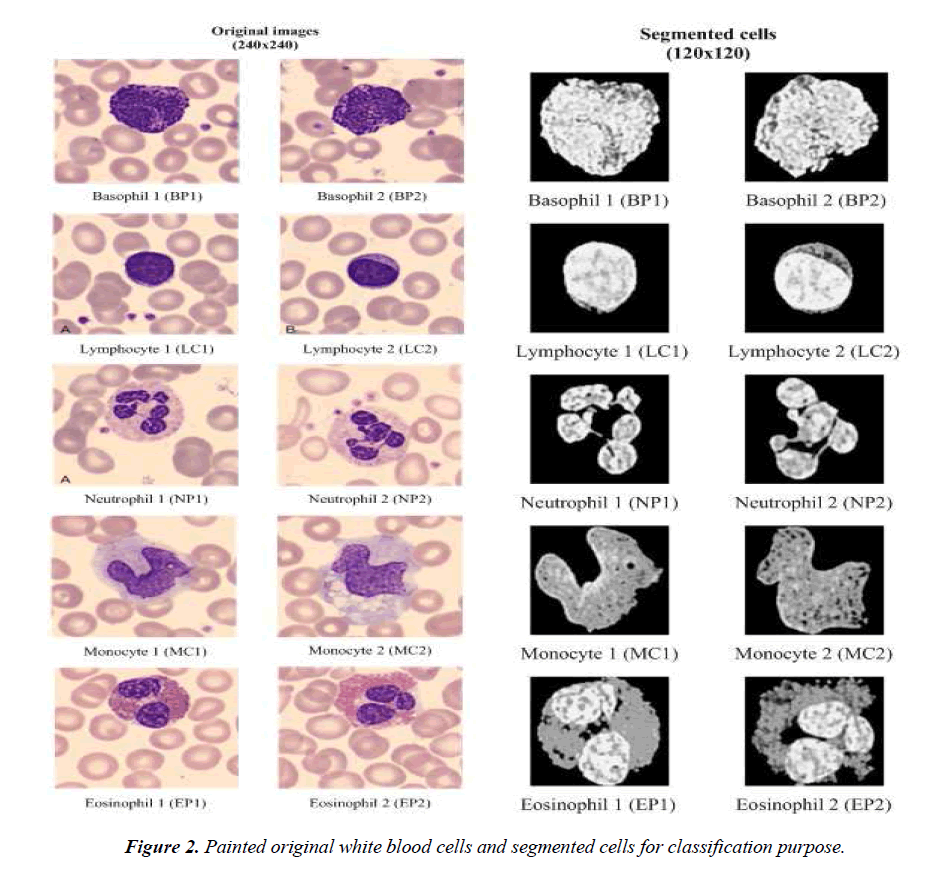
The classification process gives an output in one of the following cell types: Basophil (B), Lymphocyte (L), Neutrophil (N), Monocyte (M), or Eosinophil (E). We use two different approaches for classification, namely, SURF description and artificial neural network structures in order to enforce the results obtained. Matching capability of the SURF description is used as classifier purpose. In this approach, the images of cells that are obtained after cell extraction by image processing module are matched against a known image. Neural network structures are classifiers based on training samples. Examples of painted original white blood cells are given in Fig.2. They are typical examples of white blood cells and put here for illustration purposes of the method. The segmented cells are also given in Fig.2. The inputs of the classifiers are the segmented cells.
Surf
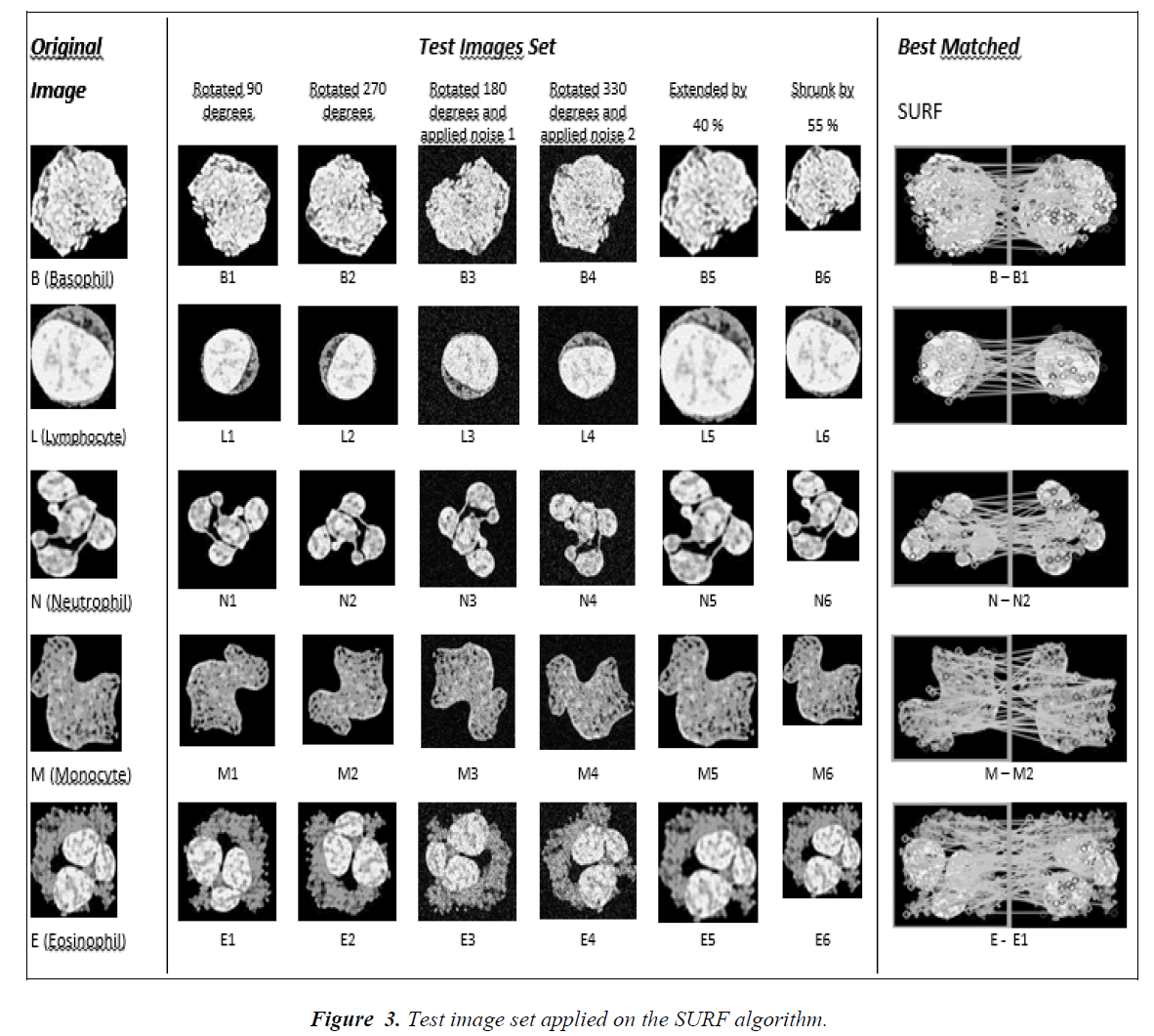
The first method used for the cell classification is Speeded-Up Robust Features (SURF) image matching scheme. The algorithm is first suggested by Bay et. al. [19] and it is based on the commonly known SIFT (Scale- Invariant Feature Transform), first suggested by Lowe [20]. The algorithm is implemented in three stages: detection of feature points, description of feature points and matching of the points. The algorithm uses the 2D Haar wavelet responses and integral images in order to get the feature points. It uses an approximation to the determinant of Hessian blob detector, For features, it uses the sum of the Haar wavelet response around the point of interest. In order to apply SURF image matching scheme to cell classification, we first detected the SURF feature points of the original cell images (L (Lymphocyte), N (Neutrophil), E (Eosinophil), M (Monocyte), and B (Basophil)). Secondly, in order to obtain a test image set, we modified the original images as follows: rotated by 90 degrees, rotated by 270 degrees, rotated 180 degrees and added Gaussian white noise having zero mean and variances of 0.01, rotated 330 degrees and added Gaussian white noise having zero mean and variances of 0.025, and finally extended by 40% and shrunk by 55% (Fig.3). After obtaining the test images, we calculated the SURF feature points of the test images. The feature points of the original images and test images are compared and the matching points are calculated by the Nearest Neighborhood principle. Based on the number of matching points the cell classification is performed (i.e. the image belongs to the class in which the number of matching feature points is maximum).
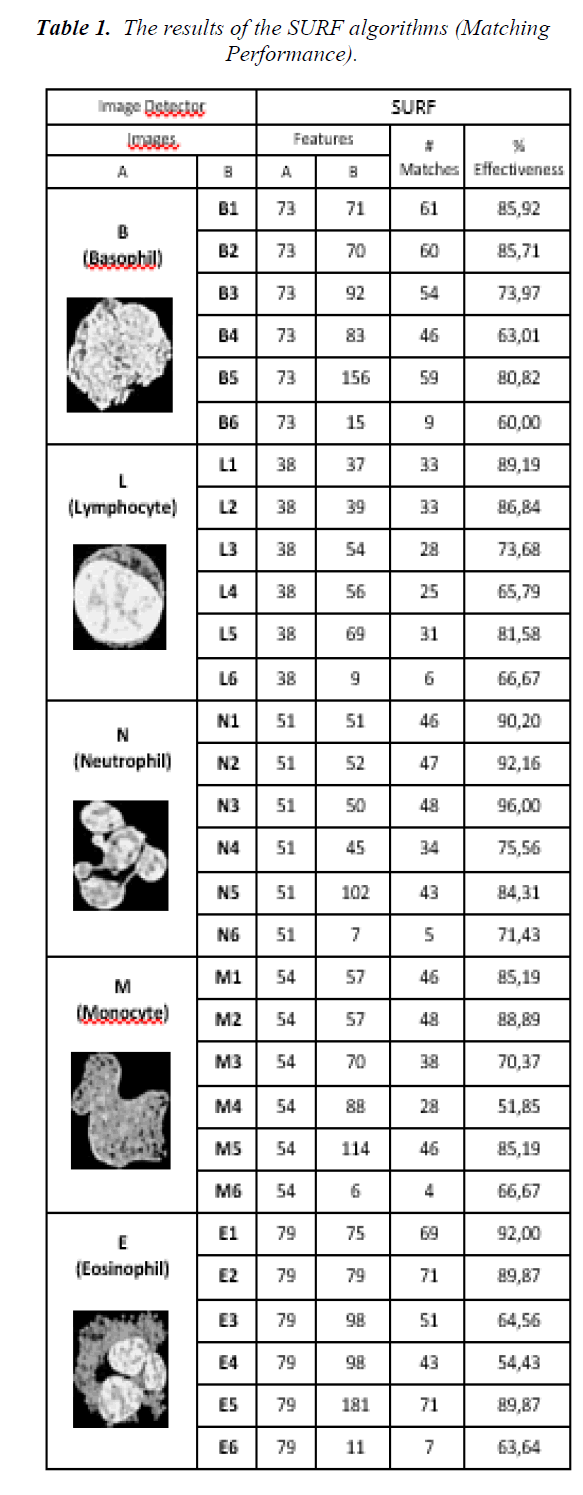
The results of the SURF classifications are given in the Table 1. In the table, the original cell images, the number of features of the images, the number of matching features, effectiveness as a percentage and an example of feature matching plot is given. The effectiveness is calculated as the ratio of the number of matches and the minimum of number of feature points of A and B columns. Here, the column A represents the number of feature points of the original images, the column B represents the number of feature points of segmented cells at the middle of the table (X1, X2, X3, X4, X5 and X6 where X represents L, N, E, M, or B). For example, the effectiveness of L and L1 is 33/38=89.19% and E and E2 is 71/79=89.87%. The number of effectiveness varies from 52% to 96% depending on the images. Average effectiveness of the process is 77.20%. Among them, 16 out of 30 are greater than 80%, and 12 out of 30 are between %60 and %80. This means that the effectiveness of the classification process by SURF can be considered as greater than 50%.
PCA, Hierarchical clustering and Dendogram
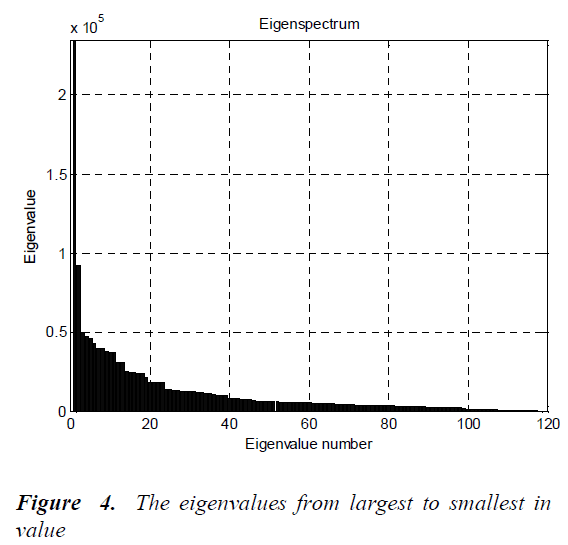
The BP1, BP2, LC1, LC2, NP1, NP2, MC1, MC2, EP1 and EP2 are rotated by the steps of 30 degrees in a counterclockwise direction around their center points. Thus, the number of images for each cell type is 24. The Principal Components Analysis (PCA) is applied to all images (120 images, in total) for dimension reduction. After this analysis, the eigenvalues from largest to smallest in value are given in Fig.4.
The percentage of the variance change is computed by using the following equation,
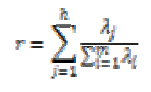
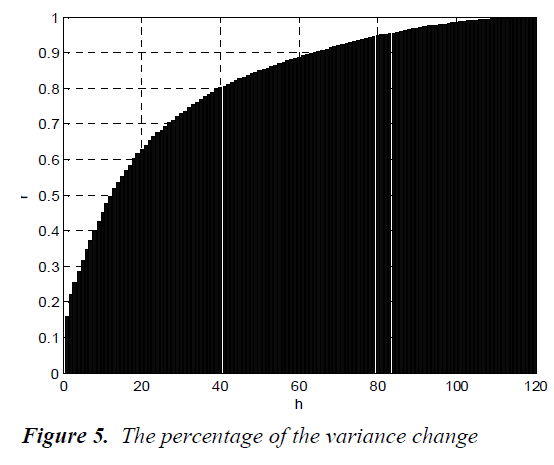
where λjs are the eigenvalues of the data set, m is the number of eigenvalues and h is the dimension number of new data set after dimension reduction (h<m). The percentage of the variance change is given in Fig.5.
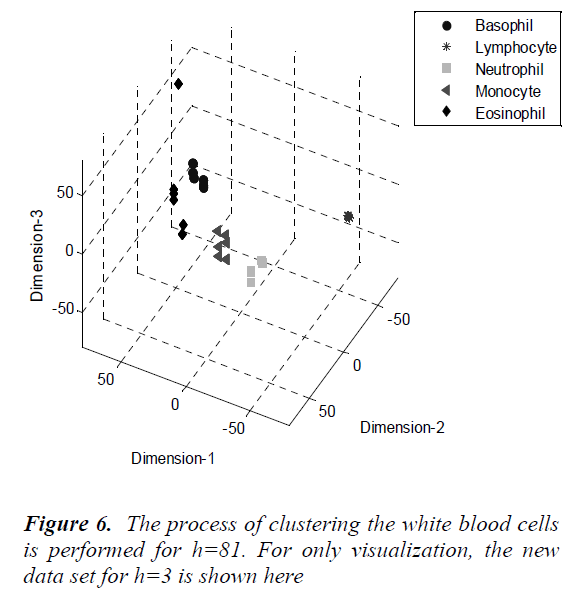
As seen from the figure, the percentage of the variance change is 95% for h=81. The process of clustering the white blood cells is performed for h=81. For only visualization, the new data set for h=3 is shown in Fig.6.
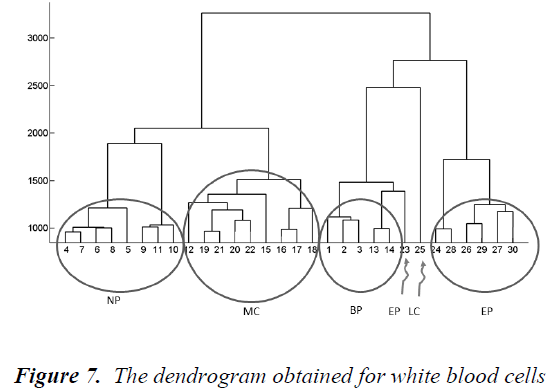
Hierarchical clustering is a method that combines data points into clusters, those clusters into larger clusters, and so forth, creating a hierarchy. A tree representing this hierarchy of clusters is known as a dendrogram. Individual data objects are the leaves of the tree, and the interior nodes are nonempty clusters [21]. The dendrogram obtained for white blood cells is shown in Fig.7.
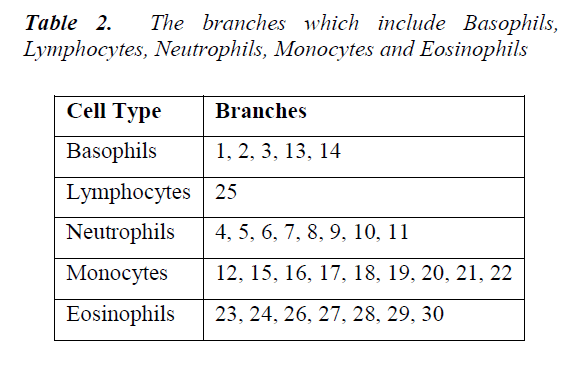
Firstly, city block distance is used to compute inter-sample dissimilarities. Then, Ward's method is preferred to calculate the dissimilarity between the merged points and the other samples. As seen from the dendrogram, the branches are enumerated. In Table 2, the branches which include Basophils, Lymphocytes, Neutrophils, Monocytes and Eosinophils are given. A different image including a white blood cell can be classified by calculating its distance from the branches in this dendrogram. It belongs to the closest branch. Although the branch number 23 belongs to an upper branch of BP, any image resembling for example a cell of EP can fall close to the branch 23. That is, classification works well enough.
Conclusion
In this work, a new automatic system used to help the diagnosis of some important blood diseases is developed, tested and the results are presented. Two types of classifiers are tried in the system. The most important result is that the effectiveness of the classification process by SURF can be considered as greater than 50%. The SURF feature points of the original cell images are detected. The number of effectiveness varies from 52% to 96% depending on the images.
Average effectiveness of the process is 77.20.
The Principal Components Analysis (PCA) is applied to the images in the training and test sets. PCA is applied to all images (120 images, in total) for dimension reduction. We choose h = 81 since r(i) = 95% is firstly achieved by 81nd eigenvector. Therefore, each image can be represented with 81 variables instead of 14400 (120x120). However, we can visualize the new data set derived by using h = 3. It is noted that the classification can be achieved clearly. In the dendrogram, the branches are enumerated. The branches which include Basophils, Lymphocytes, Neutrophils, Monocytes and Eosinophils are given. Classifications can be achieved by observing closest branch.
References
- Dorini LB, Minetto R, Leite NJ. Semiautomatic white blood cell segmentation based on multiscale analysis. IEEE J of Biomedical and Health Informatics 2013; 17 (1): 250-256. http://dx.doi.org/10.1109/TITB.2012.2207398
- Lii Q, Wang YH. Liu H, Wang J, Guo F. A combined spatial-spectral method for automatic white blood cells segmentation. Elsevier, Optics & Laser Technology, 54 (2013) 225-231. http://dx.doi.org/10.1016/j.optlastec.2013.05.022
- Nazlibilek, S, Karacor D, Ercan T, Sazli MH, Kalender O, Ege Y. Automatic segmentation, counting, size determination and classification of white blood cells. Elsevier, Measurement, 55 (2014), 58-65. http://dx.doi.org/10.1016/j.measurement.2014.04.008
- Kalina J. Classification methods for highdimensional genetic data. Elsevier, Biocybernetics and biomedical engineering, 34 (2014), 10-18. http://dx.doi.org/10. 1016/j.bbe. 2013.09.007
- Vanden Branden K, Hubert M. Robust classification in high dimensions based on the SIMCA method. Elsevier, Chemometrics and Intelligent Laboratory Systems, 79 (2005) 10-21. http://dx.doi.org/10.1016/j. chemolab.2005.03.002
- Su MC, Cheng CY, Wang PC. A Neural-Network based approach to white blood cell classification. Hindawi, The Scientific World Journal, Vol. 2014, Article ID 796371, 9 pages, http://dx.doi.org/10.1155/ 2014/796371
- Tabrizi PR, Rezatofighi SH. Yazdanpanah MJ. Using PCA and LVQ Neural Network for Automatic Recognition of Five Types of White Blood Cells. 32nd Annual International Conference of the IEEE EMBS Buenos Aires, Argentina, Aug 31-Sep4, 2010. http://dx.doi.org/10.1109/IEMBS.2010.5626788
- Apostolopulos G, Tsinopoulos SV, Dermatas E. A methodology for estimating the shape of biconcave red blood cells using multicolor scattering images. Elsevier, Biomedical Signal Processing and Control 8 (2013) 263-272. http://dx.doi.org/10.1016/j.bspc. 2012.11.002
- Arslan S, Ozyurek E, Demir CG. A color and shape based algorithm for segmentation of white blood cells in peripheral blood and bone marrow images. Cytometry Part A, Vol.85A, Issue:6, Pages 480-490, Jun 2014. http://dx.doi.org/10.1002/cyto.a.22457
- Theera-Umpon, N. Patch-based white blood cell nucleus segmentation using fuzzy clustering. ECTI Transactions on Electrical Engineering, Electronics, and Communications, Vol. 3, No. 1, 2005, pp. 15-19.
- Madhloom HT, Kareem SA, Ariffin H, Zaidan AA, Alanazi HO and Zaidan BB. An automated white blood cell nucleus localization and segmentation using image arithmetic and automatic threshold. Journal of Applied Sciences, Vol. 10, Issue 11, 2010, pp. 959-966. http://dx.doi.org/10.3923/jas.2010.959.966
- Scotti F. Robust Segmentation and Measurement Techniques of White Cells in Blood Microscope Images. IMTC 2006-Instrumentation and Measurement Technology Conference, Sorrento, Italy, 24-27 April 2006, pp. 43-48. http://dx.doi.org/10.1109/IMTC.2006. 328170
- Scotti F. Automatic Morphological Analysis for Acute Leukemia Identification in Peripheral Blood Microscope Images. in IEEE International Conference on Computational Intelligence for Measurement Systems and Applications, 2005, pp. 96–101. http://dx.doi.org/10.1109/CIMSA.2005.1522835
- Huang, DC, Hung, KD, Chan YK. A computer assisted method for leukocyte nucleus segmentation and recognition in blood smear images. The Journal of Systems and Software, Elsevier Science Publication, Vol. 85, Issue 9, September, 2012, pp. 2104-2118. http://dx.doi.org/10.1016/j.jss.2012.04.012
- Eom S, Kim S, Shin V, et al. Leukocyte Segmentation in Blood Smear Images Using Region-Based Active Contours. Advanced Concepts for Intelligent Vision Systems, Lecture Notes in Computer Science, pp. 867-876: Springer Berlin/Heidelberg, 2006. http://dx.doi. org/10.1007/11864349_79
- Mohapatra S, Patra D, and Kumar K. Blood microscopic image segmentation using rough sets. Image Information Processing (ICIIP), 2011 International Conference on, 2011, pp. 1-6. http://dx.doi.org/10.1109/ICIIP.2011.6108977
- Hiremath PS, Bannigidad P, Geeta S. Automated Identification and Classification of White Blood Cells (Leukocytes) in Digital Microscopic Images. 2010.
- Mirčić S, Jorgovanović N. Automatic Classification of Leukocytes. Journal of Automatic Control, 2006.
- Bay H, Ess A, Tuytelaars T, Van Gool L. Speeded-up robust features (SURF). Computer Vision ECCV 2006, Vol. 3951. Lecture Notes in Computer Science. p. 404-417. http://dx.doi.org/10.1007/11744023-32
- Lowe, D.G. Distintive image feature from scaleinvariant keypoints. International Journal of Computer Vision 2004; 60 (2): 91-110. http://dx.doi.org/10. 1023/B%3AVISI.0000029664.99615.94
- Kogan, J, Nicholas, C. Teboulle, M. Grouping Multidimensional Data-Recent Advances in Clustering. Springer-Verlag Berlin Heidelberg, 268, (2006) The Netherlands.