ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 16
Transcription of synaptic plasticity-related genes in patients with somnipathy combined with type 2 diabetes
Yi Zhang1, Rui Ma1, Shaohong Zou1, Gaiyu Tong1, Gulibakeranmu Abula1, Manna Hu1 and Qing Dai2*
1Department of Clinical Psychology, People’s Hospital of Xinjiang Autonomous Region, Urumchi, Xinjiang, PR China
2Department of Anesthesiology, the First Affiliated Hospital of Xinjiang Medical University, Urumchi, Xinjiang, PR China
- *Corresponding Author:
- Qing Dai
Department of Anesthesiology
The First Affiliated Hospital of Xinjiang Medical University
Urumchi, Xinjiang, PR China
Accepted on July 12, 2017
Objective: To investigate DNA methylation and hydroxymethylation in patients with somnipathy combined with type 2 diabetes, and to explore the effects of DNA methylation and hydroxymethylation on gene expression.
Methods: Thirty patients with somnipathy combined with type 2 diabetes and 20 patients with type-2 diabetes but without somnipathy were considered. DNA methylation of Disks Large Homolog 4 (DLG4) and Ras-related protein Rab-11 (Rab11) was detected by bisulfite sequencing and DNA hydroxymethylation of activity-regulated cytoskeleton-associated protein (Arc), Cyclic AMP-Responsive Element-Binding protein 3 (CREB3) and Early Growth Response protein 1 (EGR1) was analyzed by CHIP analysis. Transcription levels of DLG4, Rab11, Arc, CREB3 and EGR1 were detected by quantitative real-time RT-PCR (qRT-PCR).
Results: Methylation levels of DLG4 and Rab11 and hydroxymethylation levels of Arc, Creb3 and Erg1 in patients with somnipathy were significantly higher than those in control group (p<0.01). Increased transcription levels of DLG4, Arc and Erg1, and decreased transcription levels of Rab11 and Creb3 were found in patients with somnipathy than in patients without somnipathy. Transcription level of DLG4 was positively, and Rab11 was negatively correlated with their methylation levels. Transcription levels of Arc and Erg1 were positively, and transcription level of Creb3 was negatively correlated with hydroxymethylation levels.
Conclusion: Increased methylation levels of DLG4 and Rab11 and hydroxymethylation levels of Arc, Creb3 and Erg1 were related to the development of type 2 diabetes in patients with somnipathy. Methylation and hydroxymethylation can significantly affect gene expression at transcription level.
Keywords
Type 2 diabetes, Somnipathy, DNA methylation, Bisulfite sequencing.
Introduction
Somnipathy refers to the abnormal sleep patterns that can seriously affect normal, mental, emotional and physical functioning of patients [1]. Somnipathy can affect both children and adults [2,3]. According to 2014 report of National Sleep Foundation, the average sleeping time of children between 6-17 y is 1-2 h shorter than the recommended, resulting in emotional and physical problems. An investigation on workers has shown that, chronic sleep deprivation caused by long work hours was found to be closely correlated poor work outcomes including occupational accidents presenteeism and absenteeism [4]. Numerous clinical studies have shown that poor sleep quality and chronic sleep disorders can contribute to the onset, development and progression of metabolic diseases including insulin resistance and obesity [5-7]. Sleep disorders are usually accompanied by the emergence of endocrine disorders and inflammatory response, which may eventually lead to the occurrence of type 2 diabetes but not type 1 [8-12]. It’s well accepted that DNA methylation and hydroxymethylation are closely correlated with the prevalence of type 2 diabetes, and the control of DNA methylation may provide new opportunities for the prevention of this disease [13,14]. Sleep disorder can also be significantly affected by epigenetic factors. It has been proved that that epigenetic factors could regulate the expression of genes involved in circadian clock networks at both transcriptional and post-transcriptional levels [15]. As an important component of circadian clock system, sleep plays pivotal roles in regulating fundamental biological rhythms controlled by circadian clock. In addition, sleep disorder can regulate gene expression in mouse brain through DNA methylation and hydroxymethylation to affect stress response and circadian rhythm [16]. In view of the pathogenesis of type 2 diabetes and the effects of sleep disorder on DNA methylation and hydroxymethylation, it will be reasonable to hypothesize that sleep disorder can also affect the development of type 2 diabetes by regulating DNA methylation and hydroxymethylation.
In this study, methylation and hydroxymethylation of synaptic plasticity-related genes were detected in type 2 diabetes patients with or without somnipathy. Correlations between gene transcription, methylation, and hydroxymethylation were also analyzed. The report is as follow.
Materials and Methods
Objects
Thirty patients with somnipathy combined with type 2 diabetes were selected in the First Affiliated Hospital of Xinjiang Medical University from January 2015-2016 according the criteria described in ICSD-3.
Inclusion criteria: (1) With complaints of insomnia, including difficulties in falling asleep or sleep maintenance and early morning awakenings; (2) With daytime functional impairment, including fatigue, emotional problems and lack of motivation; (3) With symptoms cannot be explained by the lack of sufficient opportunity or suitable environment; (4) With frequency of sleep disorder greater than three times per week and duration longer than three months.
Exclusion criteria: (1) Patients with sufficient sleeping opportunities; (2) Patients with trauma, severe liver and kidney disease, coronary heart disease, high blood pressure, blood disease, cancer and autoimmune diseases such as rheumatoid arthritis, systemic lupus erythematous etc. Among the 30 patients, there were 20 males and 10 females with an average age of 45 ± 8.32 y. 20 type 2 diabetes patients without somnipathy were selected at the same time to serve as control group. Patients in control group included 11 males and 9 females with an average age of 47 ± 6.94 y. No significant differences in age, gender, Body Mass Index (BMI) and other basic information were found between those two groups.
DNA and RNA extraction
Blood was extracted from each patient. Genomic DNA and total RNA were extracted from blood samples using a blood DNA extraction kit (No. 51104, Qiagen, Hilden, Germany) and a blood RNA extraction kit (No. 51504, Qiagen, Hilden, Germany), respectively. All operations were performed in strict accordance with the instructions. DNA and RNA samples were quantified using NanoDropTM 2000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA).
Detection of DNA methylation
DNA methylation was detected using EZ DNA Methylation- GoldTM Kit (Zymo Research, Irvine, CA, USA) according to the principle of bisulfite sequencing. After bisulfite modification, PCR reaction was performed using the modified DNA as template. PCR products were sequenced to identify methylation sites by comparing to the original sequences. Sequences of all primers (Sangon, Shanghai, China) used in PCR reaction were listed in Table 1.
| Genes | Primers | Sequences |
|---|---|---|
| DLG4 | Forward | TGGTAGGGAATATGTGTGTTT |
| Reverse | ACCTAAACTCTCCTTAAAAACTCT | |
| Forward | AGTTTTTTTTTGGGGAGGAAAGAG | |
| Reverse | ACCCCCTAAAATAATCCCTTTATAC | |
| Rab11 | Forward | ATCACCCTCTCAGGAAAGTCTAAG |
| Reverse | CTTCGAGAAATGTGCCATAATCTGAC | |
| Forward | ATTTAGGGATTTTAATTAGAGATTT | |
| Reverse | CCTAACTCTACAATTCCAAAAAAAC | |
| Arc | Forward | GGTTAATGGGAGTTAGGGTTT |
| Reverse | TCATATAATCCAACTCCATCTACTC | |
| Forward | TGTGATTTTGTAGATTGGTAAGTGT | |
| Reverse | CAAACCTTAATAAACTTCTTCCAAC | |
| Creb3 | Forward | TGAGGGAATGCGATAAGTTGC |
| Reverse | TGGTCCAGTTCGTCTGAGTATT | |
| Forward | TTTCAGTGTGAACCATCCGAG | |
| Reverse | AGCACCATTCTTGAGAACTCAG | |
| Erg1 | Forward | GATCCGAGGAGCACAGAC |
| Reverse | GTGAGCCGAGCAGAAAAC | |
| Forward | GCGAAGCCCCTAAGACAAT | |
| Reverse | CACCCCAGATGAAATCCCTAC |
Table 1. Sequences of primers used in PCR reactions.
Detection of DNA hydroxymethylation
Genomic DNA (3 μg) was broken into small fragments using ultrasonic waves, and fragments ranged from 200 to 500 bp were harvested using ZymocleanTM Gel DNA Recovery Kit (Zymo Research, Irvine, CA, USA). DNA fragments were then subjected to CHIP reaction using hMeDIP kit (Diagenode, Beijing, China) to analyze DNA hydroxymethylation.
Quantitative real-time RT-PCR (qRT-PCR)
Oligo (dT) 15 primer (Sangon, Shanghai, China) and AMV reverse transcriptase (Thermo Fisher Scientific, Waltham, MA, USA) were used to synthesize cDNA through reverse transcription using RNA as template. SYBR Primemix Ex TaqTM kit (Takata, Dalian, China) and the synthesized cDNA were used to prepare PCR reaction system. Following primers were used in PCR reaction: DLG4, qHsaCEP0051707 (Bio- Rad, Hercules, CA, USA); Rab11, qHsaCID0010203 (Bio- Rad, Hercules, CA, USA); Arc, qHsaCED0048333 (Bio-Rad, USA); Creb3, qHsaCID0016155 (Bio-Rad, Hercules, CA, USA); Erg1, qHsaCID000773 (Bio-Rad, Hercules, CA, USA); GAPDH sense primer: 5'-GAGTCAACGGATTTGGTCGT-3' and GAPDH antisense primer: 5'- TTGATTTTGGAGGGATCTCG-3'. PCR reaction conditions were: 94°C for 30 s, followed by 40 cycles of 94°C for 5 s and 55°C for 45 s. Data were processed using 2-ΔΔCt method. Relative expression level of each gene was normalized to endogenous control GAPDH.
Statistical analysis
SPSS19.0 (SPSS Inc., Chicago, IL, USA) software was used to analyze the data. Comparisons between groups were performed by t-test. p<0.05 was considered to be statistically significant.
Results
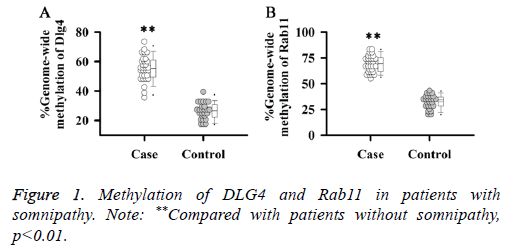
Methylation of DLG4 and Rab11
Studies have shown that DNA methylation could significantly affect the expression of DLG4, Rab11 and other genes in brain tissue of mice. Therefore, methylation levels of DLG4 and Rab11 in patients with somnipathy were investigated in this study. Results showed that methylation levels of DLG4 (Figure 1A) and Rab11 (Figure 1B) in patients with somnipathy were significantly higher than those in control group (p<0.01).
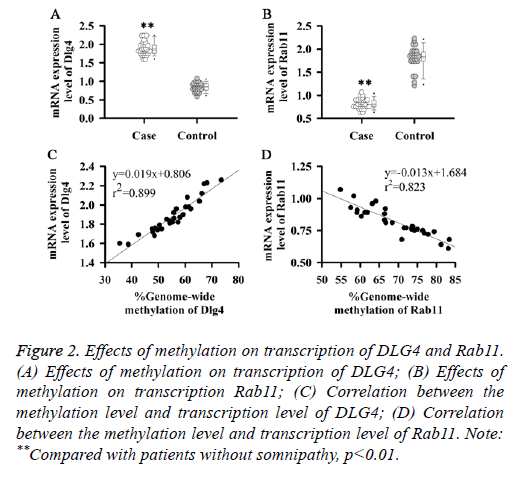
Effect of methylation on transcription of DLG4 and Rab11
DNA methylation is a common mechanism of transcriptional regulation. Methylation of the upstream transcriptional regulation region can often lead to gene silencing, while methylations within genes affect the process of transcriptional regulation or alternative splicing. Therefore, expression levels of DLG4 and Rab11 mRNA in serum of patients with somnipathy were detected in this study. As shown in Figure 2A, expression level of DLG4 mRNA in patients with somnipathy was significantly higher than that in control group (p<0.01) and was positively correlated with its methylation level (Figure 2C). While expression level of Rab11 mRNA in patients with somnipathy (Figure 2B) was significantly lower than that in control group (p<0.01) and was negatively correlated with its methylation level (Figure 2D). Those results suggested that DNA methylation could up-regulate the transcription of DLG4 but down-regulate the transcription of Rab11.
Figure 2: Effects of methylation on transcription of DLG4 and Rab11. (A) Effects of methylation on transcription of DLG4; (B) Effects of methylation on transcription Rab11; (C) Correlation between the methylation level and transcription level of DLG4; (D) Correlation between the methylation level and transcription level of Rab11. Note: **Compared with patients without somnipathy, p<0.01.
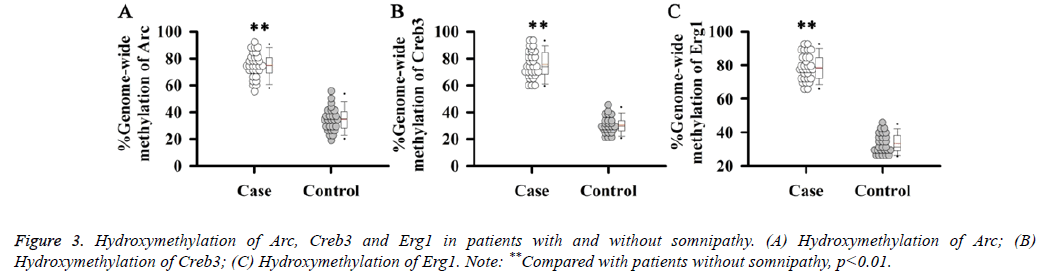
Hydroxymethylation of Arc, Creb3 and Erg1
In addition to methylation, hydroxymethylation also plays an important role in the regulation of gene expression. Therefore, hydroxymethylation levels of Arc, Creb3 and Erg1 genes were detected. Results showed that the hydroxymethylation levels of Arc (Figure 3A), Creb3 (Figure 3B) and Erg1 (Figure 3C) were significantly increased in patients with somnipathy compared with patients without somnipathy (p<0.01).
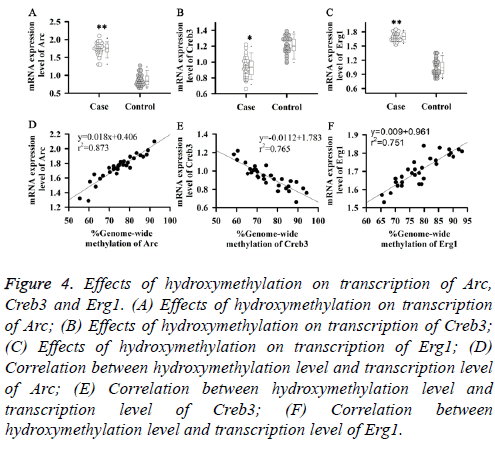
Effects of hydroxymethylation on transcription of Arc, Creb3 and Erg1
DNA hydroxymethylation plays an important role in regulating gene expression. Therefore, effects of hydroxymethylation on transcription levels of Arc, Creb3 and Erg1 were investigated in this study. Results showed that transcription levels of Arc and Erg1 were significantly increased in patients with somnipathy (Figures 4A and 4C), and were positively correlated with their hydroxymethylation levels (Figures 4D and 4F). While transcription level of Creb3 was significantly decreased in patients with somnipathy (Figure 4B), and was negatively correlated with its hydroxymethylation level (Figure 4E). Those results suggested that hydroxymethylation could up-regulate the transcription levels of Arc and Erg1, but downregulate the transcription level of Creb3.
Figure 4: Effects of hydroxymethylation on transcription of Arc, Creb3 and Erg1. (A) Effects of hydroxymethylation on transcription of Arc; (B) Effects of hydroxymethylation on transcription of Creb3; (C) Effects of hydroxymethylation on transcription of Erg1; (D) Correlation between hydroxymethylation level and transcription level of Arc; (E) Correlation between hydroxymethylation level and transcription level of Creb3; (F) Correlation between hydroxymethylation level and transcription level of Erg1.
Discussion
Sleep disorder is closely related to the development of various human diseases. In the study of Parkinson disease, Friederike et al. reported that Rapid Eye Movement (REM) sleep behaviour disorder, which is associated with aging, was a frequent disturbance for all stages of PD without preferred PD subtypes and gender [17]. In another study, fatigue caused by poor sleep was proved to be one of the main causes of inflammatory bowel disease [18]. Interactions between immune system and brain were mediated by a complex network formed by cytokines, endocrine hormones and autonomic nerves, while sleep disorders can disturb the function of this network to increase the levels of inflammatory mediators to participate in the progression of cardiovascular diseases [19]. Sleep controls daily physiological patterns which is important for normal metabolic health. Sleep deficiencies caused by circadian misalignment, night eating syndrome, shift work, sleep apnea, insufficient sleep schedules, narcolepsy and many other factors can potentially cause metabolic dysregulation, leading to the occurrence of various metabolic diseases including type 2 diabetes, obesity etc., [20]. Numerous studies have shown that the reduced sleep quality could contribute to the development of type 2 diabetes [21]. A recent large-scale clinical study has shown that patients with sleep insufficiency was more likely to develop type 2 diabetes compared with the ones with sufficient sleep and quality and quantity of sleep can be used to predict the risk of the progression of type 2 diabetes [22]. In clinical treatment of type 2 diabetes, insufficient sleep was found to be significantly related to the poor treatment outcomes of blood glucose control [23]. Besides insufficient sleep, long sleep was also found to be positively correlated with the development of various diseases including diabetes, obesity, cardiovascular diseases and hypertension, and sleep duration of 7-8 h can directly and indirectly decrease the risk of chronic disease [24].
It has been shown that methylation plays an important role in the occurrence, development, and progression of sleep disorder-mediated diseases. As a regulator of T regulatory lymphocytes, methylation level of FOXP3 gene was found to be significantly increased in children with obstructive sleep apnea, and the increased methylation level of FOXP3 can eventually lead to the emergence of inflammatory responses [25]. Therefore, methylation level of FOXP3 can potentially be a biomarker for the vulnerability of organs. A recent study has shown that wakefulness in a single night is sufficient to alter the transcriptional profiles of genes involved in circadian clock system in key metabolic tissues through DNA methylation, which can be used to explain the disrupted metabolic integrity caused by shift work [26]. As a member of Membrane- Associated Guanylate Kinase (MAGUK) family, DLG4 is an innate immune receptor that plays pivotal roles in synaptic plasticity [27]. Rab11 controls the transportation and signal transduction of DLG4 [28]. It has been widely accepted that synaptic plasticity is closely related to sleep status [29]. A previous study has shown that methylation level of DLG4 and Rab11 were altered in mice with sleep disorders, and the changed methylation levels of DLG4 and Rab11 were proved to be related to the changes in stress response and circadian rhythm [14]. In our study, methylation levels of DLG4 and Rab11 were found to be significantly increased in type 2 diabetes patients combined with somnipathy compared with patients without somnipathy. In addition, transcription level of DLG4 was positively related to methylation level, and transcription level of Rab11 was negatively related to methylation level. Those results suggested that changes in transcription profile of DLG4 and Rab11 caused by methylation were related to the development of type 2 diabetes in patients with somnipathy, and different methylation patterns may regulate transcription to different directions.
DNA hydroxymethylation also play pivotal roles in regulating genome-wide gene transcription pattern [30]. Arc, Creb3 and Erg1 are synaptic plasticity genes, and hydroxymethylation levels of Arc, Creb3 and Erg1 were found to be significantly altered in mice after sleep deprivation [16]. Consistent with previous studies, in our study, hydroxymethylation levels of Arc, Creb3 and Erg1 in patients with somnipathy combined with type 2 diabetes were found to be significantly higher than those in patients without somnipathy. In addition, transcription levels of Arc and Erg1 were found to be positively correlated with their hydroxymethylation levels, while transcription level of Creb3 was found to be negatively correlated with its hydroxymethylation level. Those results suggested that changes in transcription profiles of Arc, Creb3 and Erg1 caused by DNA hydroxymethylation were related to the development of type 2 diabetes in patients with somnipathy, and the different directions of transcription regulation may be explained by different patterns of DNA hydroxymethylation.
In conclusion, increased methylation levels of DLG4 and Rab11, and hydroxymethylation levels of Arc, Creb3 and Erg1, which significantly affected gene expression, were related to the development of type 2 diabetes in patients with somnipathy.
References
- Zhao ZX. Science of clinical somnipathy. Second Military College Press 2003.
- Knutson KL, Van Cauter E, Rathouz PJ. Trends in the prevalence of short sleepers in the USA: 1975-2006. Sleep 2010; 33: 37-45.
- Bryson WJ, Edwards CL. A brief review of sleep normality and pathology among adult Black men. J Afr Am Studies 2013; 17: 426-432.
- Swanson LM, Arnedt J, Rosekind MR. Sleep disorders and work performance: findings from the 2008 National Sleep Foundation Sleep in America poll. J Sleep Res 2011; 20: 487-494.
- Androutsos O, Moschonis G, Mavrogianni C. Identification of lifestyle patterns, including sleep deprivation, associated with insulin resistance in children: the Healthy Growth Study. Eur J Clin Nutr 2014; 68: 344.
- Buxton OM, Pavlova M, Reid EW. Sleep restriction for 1 week reduces insulin sensitivity in healthy men. Diabetes 2010; 59: 2126-2133.
- Cappuccio FP, Taggart FM, Kandala NB. Meta-analysis of short sleep duration and obesity in children and adults. Sleep 2008; 31: 619-626.
- Briançon-Marjollet A, Weiszenstein M, Henri M. The impact of sleep disorders on glucose metabolism: endocrine and molecular mechanisms. Diabetol Metabol Syndrome 2015; 7: 25.
- Faraut B, Boudjeltia KZ, Vanhamme L. Immune, inflammatory and cardiovascular consequences of sleep restriction and recovery. Sleep Med Rev 2012; 16: 137-149.
- Mullington JM, Simpson NS, Meier-Ewert HK. Sleep loss and inflammation. Clin Endocrinol Metabol 2010; 24: 775-784.
- Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Invest 2011; 121: 2111-2117.
- Barone MTU, Menna-Barreto L. Diabetes and sleep: a complex cause-and-effect relationship. Diabetes Res Clin Pract 2011; 91: 129-137.
- Chambers JC, Loh M, Lehne B. Epigenome-wide association of DNA methylation markers in peripheral blood from Indian Asians and Europeans with incident type 2 diabetes: a nested case-control study. Lancet Diabetes Endocrinol 2015; 3: 526-534.
- Ling C. Epigenetic modifications and type 2 diabetes in humans. Genet Diabetes 2014; 23: 102-110.
- Qureshi IA, Mehler MF. Epigenetics of sleep and chronobiology. Curr Neurol Neurosci Rep 2014; 14: 1-11.
- Massart R, Freyburger M, Suderman M. The genome-wide landscape of DNA methylation and hydroxymethylation in response to sleep deprivation impacts on synaptic plasticity genes. Transl Psychiatr 2014; 4: e347.
- Sixel-Döring F, Trautmann E, Mollenhauer B. Associated factors for REM sleep behaviour disorder in Parkinson disease. Neurol 2011; 77: 1048-1054.
- Graff LA, Vincent N, Walker JR. A population‐based study of fatigue and sleep difficulties in inflammatory bowel disease. Inflamm Bowel Dis 2011; 17: 1882-1889.
- Motivala SJ. Sleep and inflammation: psychoneuroimmunology in the context of cardiovascular disease. Ann Behavioral Med 2011; 42: 141-152.
- Depner CM, Stothard ER, Wright Jr KP. Metabolic consequences of sleep and circadian disorders. Curr Diabetes Rep 2014; 14: 1-9.
- Rajan P, Greenberg H. Obstructive sleep apnea as a risk factor for type 2 diabetes mellitus. Nature Sci Sleep 2015; 7: 113.
- Cappuccio FP, D'elia L, Strazzullo P. Quantity and quality of sleep and incidence of type 2 diabetes. Diabetes Care 2010; 33: 414-420.
- Tsai YW, Kann NH, Tung TH. Impact of subjective sleep quality on glycemic control in type 2 diabetes mellitus. Family Pract 2012; 29: 30-35.
- Buxton OM, Marcelli E. Short and long sleep are positively associated with obesity, diabetes, hypertension, and cardiovascular disease among adults in the United States. Social Sci Med 2010; 71: 1027-1036.
- Kim J, Bhattacharjee R, Khalyfa A. DNA methylation in inflammatory genes among children with obstructive sleep apnea. Am J Respir Crit Care Med 2012; 185: 330-338.
- Cedernaes J, Osler ME, Voisin S. Acute sleep loss induces tissue-specific epigenetic and transcriptional alterations to circadian clock genes in men. J Clin Endocrinol Metab 2015; 100: 1255-1261.
- Meyer D, Bonhoeffer T, Scheuss V. Balance and stability of synaptic structures during synaptic plasticity. Neuron 2014; 82: 430-443.
- Husebye H, Aune MH, Stenvik J. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory factor-3 on phagosomes. Immunity 2010; 33: 583-596.
- Smith C. Sleep states, memory processes and synaptic plasticity. Behav Brain Res 1996; 78: 49-56.
- Wang T, Pan Q, Lin L. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum Mol Genet 2012.