ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 22
Structural changes of gut microbiota in mice with chronic constipation and intestinal tumor
1Key Laboratory for Forest Resources Conservation and Use in the Southwest Mountains of China, Ministry of Education, Southwest Forestry University, PR China
2Southwest Forestry University, PR China
- *Corresponding Author:
- Yunpeng Luan
Key Laboratory for Forest Resources Conservation and Use in the Southwest Mountains of China
Ministry of Education
Southwest Forestry University, PR China - Chengzhong He
Key Laboratory for Forest Resources Conservation and Use in the Southwest Mountains of China
Ministry of Education
Southwest Forestry University, PR China
Accepted date: December 22, 2017
Background: Constipation is a highly frequent complication in patients with chronic gastrointestinal disorder and intestinal cancer. The purpose of this study was to evaluate the changes of gut microbiota in the disease development from chronic constipation to intestinal tumor.
Methods: Loperamide-induced constipation model was established to verify that chronic constipation can increase the risk of intestinal tumor. 128 Kunming mice were divided into four groups, namely, healthy, chronic constipation, intestinal tumor induced by chronic constipation and intestinal tumor with constipation after DMH (dimethylhydrazine) induction. The fecal bacterial diversity was profiled by the V3 and V4 region of the 16S ribosomal RNA genes.
Result: Healthy mice showed enrichment in operational taxonomic units (OTUs) affiliated with members of the Lactobacillus, Clostridium XlVa, Allobaculum and etc. Chronic constipation mice showed the decreases in OTUs affiliated with members of the Lactobacillus genus and the increase of the Barnesiella genus. Tumor-bearing mice showed enrichment in OTUs affiliated with members of the Barnesiella.
Conclusions: These results suggest that changes in the gut microbiota in mice with chronic constipation probably contribute to tumorigenesis and modulation of the gut microbiota may help to alleviate chronic constipation and eventually prevent the development of colon cancer.
Keywords
Chronic constipation, Intestinal tumor, Gut microbiota, Lactobacillus, Barnesiella.
Introduction
Constipation is a worldwide functional gastrointestinal disorder that affects more than 10% of the world's population [1]. Constipation appears to be more common in the elderly, women, and impacts the quality of life in constipated people [2,3]. Accumulating evidence indicates that constipation is also significantly associated with higher prevalence and incidence of colorectal cancer and the risk increase with the severity of chronic constipation [4-6]. There is a primary mechanism of constipation, which is a failure of peristalsis to move luminal contents through colon, resulting in more time for bacterial degradation of stool solids [7]. However, the exact mechanism of chronic constipation induced colorectal cancer is still unknown.
Some significant risk factors for colorectal cancer include alcohol consumption, chronic inflammation of the gastrointestinal tract and chronic constipation [1,8]. Recently, it has been shown that many diseases are closely related to the intestinal flora [9,10]. A novel mechanism has been revealed that microbiota-induced inflammation drives colon carcinogenesis by altering at RA metabolism [11], which suggest that manipulating the microbiota may modulate cancer immunotherapy [12,13]. Selective modulation of the structure and activity of gut microbiota has been demonstrated to confer beneficial effects [14]. Therefore, evaluation for the changes of gut microbiota in the disease development from chronic constipation to intestinal tumor is a major issue to be addressed.
Loperamide acts on opioid receptors in the intestinal wall to inhibit the release of acetylcholine and prostaglandin, resulting in inhibition of intestinal peristalsis and prolonging of the retention time of intestinal contents, which lead to constipation [8]. In this study, we investigated the relation of constipation and intestinal tumor in a loperamide induced chronic constipation model, and a DMH (dimethylhydrazine) induced intestinal cancer model with chronic constipation in Kunming mice. Our work revealed chronic constipation can cause the change of intestinal microbial and increase the incidence of intestinal tumor.
Materials and Methods
Materials
A total of 128 Kunming mice were provided by Kunming Medical University. Main equipment for experiments included Leica stereomicroscope (Leica Company, Germany), optical microscope (Nikon Company, Japan), and microscopic imaging system (Nikon Company, Japan). Loperamide hydrochloride (Xian-Janssen Pharmaceutical Ltd., performance standard: YBH04562010, specifications: 2 mg/tablet, batch number: 141111266); DMH (TCI Company, Japan), and all other needed regents were analytical reagents (Xilong Chemical Co., Ltd.).
Preparation of Loperamide and DMH
For constipation induction experiments, commercial loperamide hydrochloride (2 mg/tablet) was dissolved in physiological saline to a concentration of 0.25 mg/mL, adjusted pH to 7.0, mixed well and stored at -20°C for further use. For DMH preparation, 1 g DMH hydrochloride was dissolved in 100 mL physiological saline, filtered with 0.22 um organic-phase bacteria filter, and preserved at 4°C for further use.
Experimental procedures
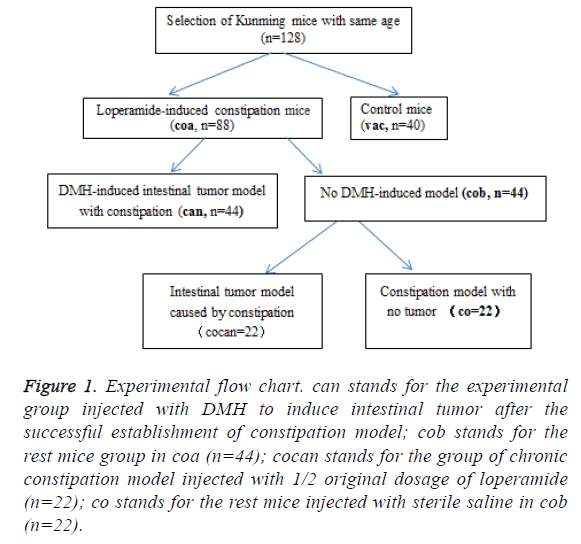
A total of 128 healthy Kunming mice with same age were screened and randomly divided into experimental group (coa, n=88) and control group (vac, n=40) (Figure 1). For induction of constipation model, the coa group was treated by intragastric administration of loperamide according to the intragastric volume per unit of body weight of 2.5 mg/(Kg*d) for 2 consecutive weeks; the vac group received intragastric administration with equal volume of sterile saline solution.
Figure 1: Experimental flow chart. can stands for the experimental group injected with DMH to induce intestinal tumor after the successful establishment of constipation model; cob stands for the rest mice group in coa (n=44); cocan stands for the group of chronic constipation model injected with 1/2 original dosage of loperamide (n=22); co stands for the rest mice injected with sterile saline in cob (n=22).
Two weeks later, the constipation model was established successfully [5].
Establishment of intestinal tumor model based on constipation model
After the successful establishment of constipation model, the coa group was randomly divided into can group and cob group. The can group was injected with DMH to induce intestinal tumor and the cob group was divided into cocan group and co group. Then the 22 mice with constipation in cob group, namely, cocan group were continuously treated with 1/2 original dosage Loperamide, which lead to chronic constipation. Stereo-microscope was used to observe lesion morphology, and lesions were fixed by 10% formalin solution and sliced into 5 μm sections according to the paraffin section method.
Based on the constipation model, DMH was injected to induce intestinal cancer model. Anatomy showed that the mice with chronic constipation had sarcoma in the intestines. Visible sarcoma under naked eyes was firstly found in the intestines of the can group at the 7th week, which was followed by the cocan group at the 9th week (Table 1). No abnormalities were detected in the vac group and co group.
| Week 5 | Week 7 | Week 9 | Week 11 | Week 13 | |
|---|---|---|---|---|---|
| can | 0 | 3 | 5 | 8 | 12 |
| cocan | 0 | 0 | 2 | 4 | 6 |
| co | 0 | 0 | 0 | 0 | 0 |
| vac | 0 | 0 | 0 | 0 | 0 |
Table 1: Changes in the number of visible sarcomas in the intestines of mice by anatomy (the number of sarcomas/mouse).
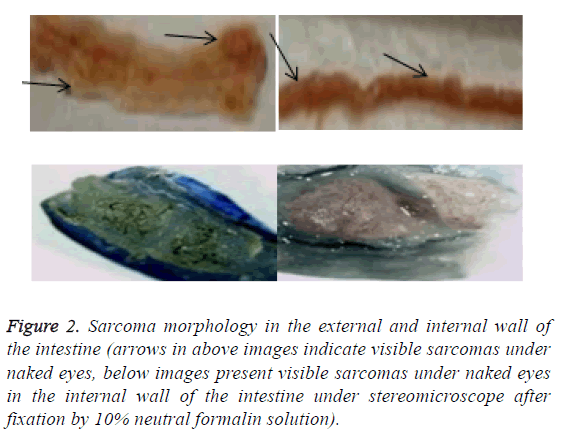
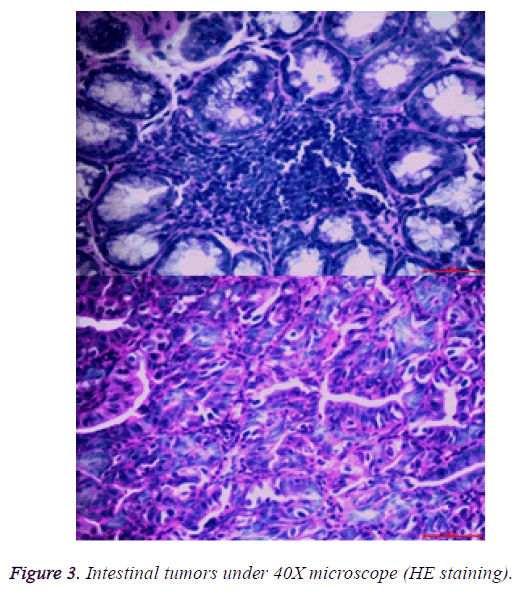
After the lesions in the intestines were fixed by 10% neutral formalin solution, sarcomas under the stereomicroscope presented uneven surface and abundant blood vessels in staggered and irregular arrangement (Figure 2). Observation of pathological sections under high-power microscope revealed that cells showed irregular shape, inconsistent size, irregular nuclear shape, nuclear hyperchromatism, prominent nucleolus and nuclear division at lesion sites, which were confirmed as tumor cells when compared with relevant data (Figure 3).
Figure 2: Sarcoma morphology in the external and internal wall of the intestine (arrows in above images indicate visible sarcomas under naked eyes, below images present visible sarcomas under naked eyes in the internal wall of the intestine under stereomicroscope after fixation by 10% neutral formalin solution).
Fecal bacterial DNA extraction
The fecal bacterial DNA of each sample was extracted according to the manufacturer’s instructions. The total DNA samples were characterized by 1% agarose gel electrophoresis for integrity and size. The DNA extracts were stored at -80°C before being used as templates for 16S rDNA analysis.
Fecal bacterial RNA extraction
The V3 and V4 region of the 16S ribosomal RNA (rRNA) gene was from each DNA sample. The RNA PCR Primer Index 47 (GAGTTCCTTGGCACCCGAGAATTCCA) was used to amplify the V3 and V4 domain of bacterial 16S rRNA. PCR reactions contained 5-100 ng DNA template, 1X GoTaq Green Master Mix, 1 mM MgCl2, and 2 pmol of each primer. Reaction conditions consisted of an initial 94°C for 3 min followed by 35 cycles of 94°C for 45 s, 50°C for 60 s, and 72°C for 90 s, and a final extension of 72°C for 10 min. All samples were amplified in triplicate and combined prior to purification. The purified libraries were sequenced on the Illumina Miseq platform.
16S rRNA gene analysis
Raw Illumina fastq files were adapter-removed using cutadapt (Marcel, EMBnetjournal), sequence-assembled by PEAR [15] and quality-controled by Prinseq [16]. 16S rRNA gene sequences were assigned to operational taxonomic unit using Usearch with a threshold of 97% pair-wise identity [17], and then classified taxonomically using the Ribosomal Database Project (RDP) classifier (http://rdp.cme.msu.edu/misc/resources.jsp), Silva (http://www.arbsilva.de/) and NCBI 16S (http://ncbi.nlm.nih.gov/).
Statistical analysis
Results are obtained by a variety of statistics and graphic software. The statistical analysis was performed with R packages and mothur [18]. Welch’s t-tests were conducted to compare the phenotypes of the different group mice and statistical significance was set at p<0.05. To determine the role of the chronic constipation and gut microbiome in the development of colon tumorigenesis, the well-established model of constipation that recapitulates the progression from chronic constipation to intestinal tumor in humans.
Results
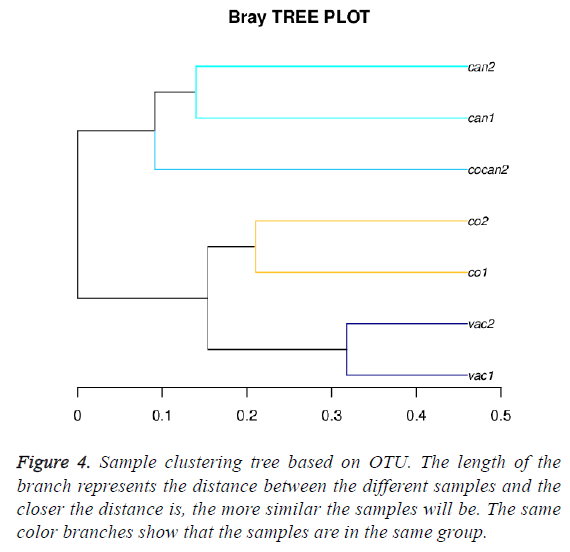
Sample clustering based on OTU abundance in different group mice
The sample clustering tree diagram can directly reflect the similarity and difference among the samples through the tree structure. According to the beta diversity distance matrix, hierarchical clustering analysis was carried out. The unweighted-pair-group method with arithmetic mean was used to construct tree structure. The tree structure could be used for visual analysis. Bray-Crutis method was used to compute distance between the different samples. The Bray-Crutis tree was consistent with our classification (Figure 4).
Variation of fecal microbial communities in different group mice
The average Shannon value of the normal mice (vac group) was significantly lower than that of the intestinal tumor mice (can and cocan group) and the average value of normal mice was slightly lower than that of the constipation mice (co group) and it was similar in the Simpson value (Table 2). These results indicated that gut microbiome diversity increase from normal to constipation and then to intestinal tumor.
| Sample ID | Seq num | OUT num | Shannon index | Coverage | Simpson | P-value |
|---|---|---|---|---|---|---|
| can1 | 45167 | 499 | 3.60 | 1.00 | 0.07 | 0.002 |
| can2 | 53559 | 527 | 4.31 | 1.00 | 0.03 | 0.050 |
| co1 | 31760 | 312 | 2.57 | 1.00 | 0.17 | 0.026 |
| co2 | 41496 | 287 | 1.88 | 1.00 | 0.30 | 0.035 |
| cocan2 | 48134 | 465 | 3.85 | 1.00 | 0.06 | 0.041 |
| vac1 | 33702 | 357 | 2.19 | 1.00 | 0.33 | 0.043 |
| vac2 | 30518 | 257 | 1.61 | 1.00 | 0.46 | 0.033 |
Table 2: Variation of fecal microbial communities in different group mice.
The changes of fecal microbial communities
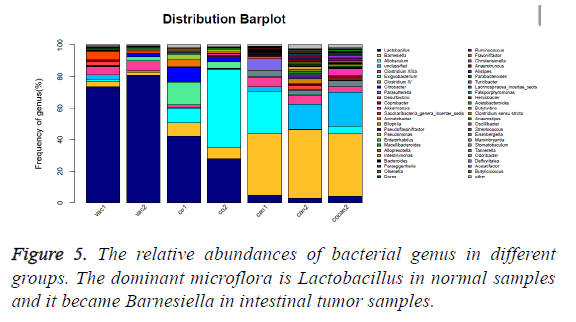
The healthy samples (vac) had higher abundance of Lactobacillus (p<0.01) and lower abundance of Barnesiella (p<0.05) than the constipation samples (co) and the intestinal tumor samples (can and cocan) (p<0.01) (Figure 5 and Table 3), suggesting that Lactobacillus is dominant microflora in healthy mice. The ratio of Lactobacillus in constipation samples (co) decreased from more than 70% to less than 45% compared to the healthy samples (vac), while that of the intestinal tumor samples (can) was less than 10%. At the same time, the ratio of Barnesiella increased from less than 5% to around 8%, while that of the intestinal tumor samples became around 40%, which indicated that the ratio of Barnesiella increased by nearly ten times in intestinal tumor mice and Barnesiella became dominant microflora.
| vac1 | vac2 | co1 | co2 | can1 | can2 | cocan2 | P-value | |
|---|---|---|---|---|---|---|---|---|
| Lactobacillus | 73.41 | 80.81 | 42.19 | 28.02 | 4.96 | 3.05 | 4.27 | 0.023 |
| Barnesiella | 3.35 | 2.05 | 8.45 | 6.96 | 38.87 | 43.34 | 39.7 | 0.025 |
| Allobaculum | 1.45 | 0 | 8.98 | 49.43 | 26.96 | 0.5 | 4.77 | 0.030 |
| unclassified | 2.95 | 1.04 | 0.9 | 0.63 | 2.8 | 15.54 | 21.54 | 0.038 |
| Clostridium XlVa | 4.96 | 6.38 | 1.68 | 0.26 | 6.03 | 5.94 | 3.1 | 0.040 |
Table 3: The relative abundances of bacterial genus in different groups.
The bacterial family composition also presented an obvious alteration in response to the development from normal mice to constipation mice and then to intestinal tumor. The bacterial family composition of normal samples mainly consisted of Lactobacillaceae, Lachnospiraceae, and Porphyromonadaceae. Erysipelotrichaceae was seldom detected in the normal samples and it grew fast in constipation and intestinal tumor samples. Ruminococcaceae was little in the normal and constipation samples and it became common in intestinal tumor samples (Table 4).
| vac1 | vac2 | co1 | co2 | can1 | can2 | cocan2 | P-value | |
|---|---|---|---|---|---|---|---|---|
| Porphyromonadaceae | 5.33 | 2.26 | 9.16 | 8.56 | 41.33 | 48.96 | 59.71 | 0.003 |
| Lactobacillaceae | 73.41 | 80.81 | 42.19 | 28.02 | 4.96 | 3.05 | 4.27 | 0.008 |
| Lachnospiraceae | 7.04 | 7.3 | 2.67 | 0.51 | 8.05 | 21.84 | 9.24 | 0.004 |
| Erysipelotrichaceae | 1.49 | 0.1 | 9.16 | 50.02 | 27.28 | 6.22 | 5.59 | 0.005 |
| Ruminococcaceae | 0.33 | 0.17 | 0.13 | 0.11 | 5.94 | 10.56 | 7.54 | 0.033 |
Table 4: The relative abundances of bacterial family in different groups.
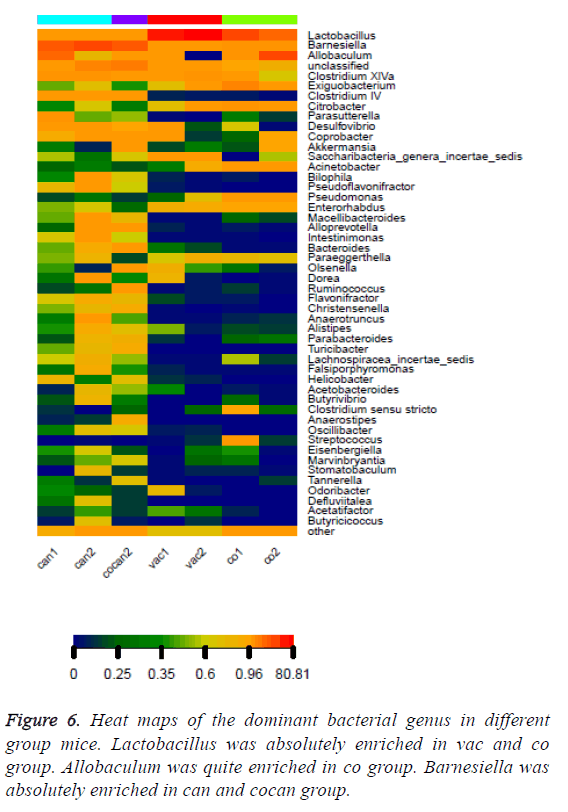
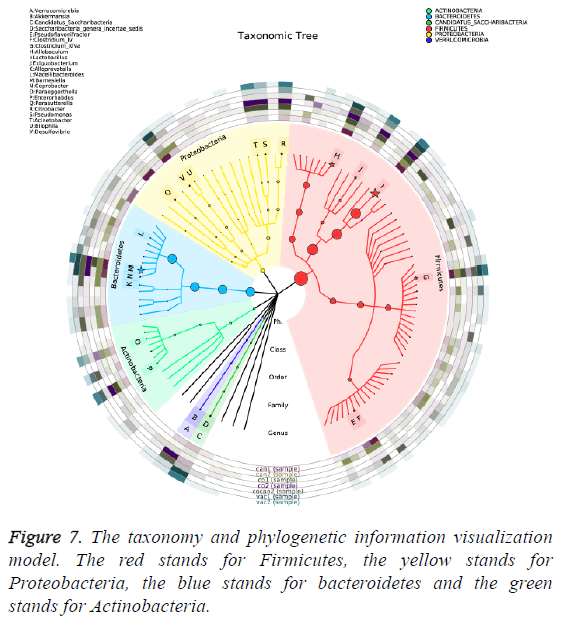
The variation of some dominant bacterial genus was presented with a heat map to figure out their contribution to the variation of the bacterial community (Figure 6). According to the results of the heat map, Lactobacillus was absolutely enriched in the normal and constipation samples. Allobaculum was quite enriched in constipation samples, and Barnesiella was absolutely enriched in intestinal tumor samples. Based on the comparison results of each sample, dominant species taxonomies were selected. Combining species abundance the taxonomy and phylogenetic information were shown in cyclic tree diagram (Figure 7).
Discussion
Accumulating evidence indicates that Gut microbiota might be associated with the etiology or development of chronic constipation and imbalances in the structure of the gut microbiota may impair gut barrier function resulting in the increase of risk for colorectal cancer development [5,6,8,10]. In this study, models of healthy, Loperamide-induced chronic constipation and DMH-induced intestinal tumor Kunming mice with chronic constipation were utilized to evaluate the structural changes of gut microbiota in the mice of chronic constipation and intestinal tumor. We showed that the abundance of Lactobacillus decreased fast in the chronic constipation and intestinal tumor samples and the abundance of Barnesiella increased slightly in the chronic constipation samples and increased fast in the intestinal tumor samples (Table 3).
Daillère et al. have reported the antitumoral efficacy of CTX (Cyclophosphamide, an immunomodulatory anticancer compound) relies on two gut commensal species, Enterococcus hirae and Barnesiella intestinihominis in a NOD2-dependent manner. These two bacteria changed the tumor microenvironment, reducing regulatory T cells and stimulating cognate antitumor CTL (cytotoxic T-lymphocyte) responses [19]. This paper showed that Barnesiella is a beneficial bacterium. In our experiment Barnesiella really took the place of Lactobacillus in gut microbiota of Kunming mice. The mechanism behind these phenomena to be studied further.
Conclusion
Through establishing a chronic constipation model in Kunming mice, it was found that chronic constipation could affect the occurrence of sarcoma in the intestines of Kunming mice. Significant changes of gut microbiota had taken place in the development from healthy to chronic constipation and then to intestinal tumor. The gut microbiome diversity also increase. Lactobacillus, a dominant microflora in normal stage was obviously inhibited in intestinal tumor stage. Meanwhile, the bacteria Barnesiella took the place of Lactobacillus and it became a new dominant microflora. The Barnesiella is probiotics and it is beneficial for human. But it really became dominant in gut microbiota in our experiments. What causes these phenomena will be further studied.
Acknowledgements
This study was funded by National Natural Science Foundation of China (NSFC) (61363061) and Advanced and Characteristic Key Biological Disciplines of Yunnan Province (project serial code: 50097505). The thesis of "Scientific and Technological Innovation Team Construction Project for Protection and Utilization of Under-forest Biological Resources (project serial code: 51400605)" was completed through joint support.
References
- Xu C, Xia Y, Huang L. Role of gut probiotic microbiota in the apoptosis of gastrointestinal cancer. Oxidation Commun 2016; 39: 214-222.
- Suares NC, Ford AC. Prevalence of, and risk factors for, chronic idiopathic constipation in the community: systematic review and meta-analysis. Am J Gastroenterol 2011; 106: 1582-1591.
- Talley NJ, Fleming KC, Evans J. Constipation in an elderly community: a study of prevalence and potential risk factors. Am J Gastroenterol 1996.
- Guérin A, Mody R, Fok B. Risk of developing colorectal cancer and benign colorectal neoplasm in patients with chronic constipation. Aliment Pharmacol Thera 2014; 40: 83-92.
- Watanabe T, Nakaya N, Kurashima K. Constipation, laxative use and risk of colorectal cancer: The Miyagi cohort study. Euro J Cancer 2004; 40: 2109-2115.
- Roberts MC, Millikan RC, Galanko JA. Constipation, laxative use, and colon cancer in a North Carolina population. Am J Gastroenterol 2003; 98: 857-864.
- Schiller L. The therapy of constipation. Alimentary Pharmacol Thera 2001; 15: 749-763.
- Zackular JP, Baxter NT, Iverson KD. The gut microbiome modulates colon tumorigenesis. MBio 2013; 4: e00692-e00713.
- Dubin K, Callahan MK, Ren B. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun 2016.
- Wang T, Cai G, Qiu Y. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISMEJ 2012; 6: 320-329.
- Bhattacharya N, Yuan R, Prestwood TR. Normalizing microbiota-induced retinoic acid deficiency stimulates protective CD8+ T cell-mediated immunity in colorectal cancer. Immunity 2016; 45: 641-655.
- Sivan A, Corrales L, Hubert N. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Sci 2015; 350: 1084-1089.
- Iida N, Dzutsev A, Stewart CA. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Sci 2013; 342: 967-970.
- Delzenne NM, Neyrinck AM, Bäckhed F. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat Rev Endocrinol 2011; 7: 639-646.
- Zhang J, Kobert K, Flouri T. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformat 2014; 30: 614-620.
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011; 27: 863-864.
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26: 2460-2461.
- Schloss PD, Westcott SL, Ryabin T. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009; 75: 7537-7541.
- Daillère R, Vétizou M, Waldschmitt N. Enterococcus hirae and barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity 2016; 45: 931-943.