ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 11
SHP-1 reverses trastuzumab resistance and migration in HER2-positive breast cancer cells through the stat3/p-stat3 pathway
Dexin Lei1,2, Rong Li2,3, Aimin Li1,2 and Rongcheng Luo1,2*
1Department of Oncology, Integrated Hospital of Traditional Chinese Medicine, Southern Medical University, Guangzhou, Guangdong 510315, PR China
2Cancer Center, Southern Medical University, Guangzhou, Guangdong 510315, PR China
3Department of Oncology, Nanfang Hospital, Southern Medical University, Guangzhou, Guangdong 510515, PR China
- *Corresponding Author:
- Rongcheng Luo
Department of Oncology
Integrated Hospital of Traditional Chinese Medicine
Southern Medical University, PR China
Accepted on April 12, 2018
DOI: 10.4066/biomedicalresearch.30-17-2421
Visit for more related articles at Biomedical ResearchDecreased expression of Src homology region 2 domain containing protein tyrosine phosphatase 1 (SHP-1) was found in human breast cancer cell lines and primary breast cancers. As a candidate tumor suppressor gene, the role of SHP-1 in HER-2-positive breast cancer remains to be elucidated. The aim of the present study was to investigate the effects of upregulated SHP-1 expression in reversing trastuzumab resistance and migration by HER-2-positive breast cancer cells, potentially providing a new target to trastuzumab resistance in anti-HER2 therapy. Western blotting analysis was used to determine the expression levels of SHP-1 in trastuzumab-resistant cells transfected with a lentivirus vector expressing GFP-(control) or GFP-SHP-1 (experimental). MTT assay was used to determine changes in sensitivity to trastuzumab between two modified cell lines after overexpressing SHP-1. Clone formation and wound-healing assays were used to detect the capacity change in cell proliferation and migration. Western blot analysis was also used to examine the expressions of c-myc, cyclinD1, p-STAT3, STAT3, and MMP-9. Results from this study indicated that SHP-1 not only reduced cell proliferation and migration, but also significantly sensitized resistant cells to trastuzumab. Data also showed that SHP-1 restores sensitivity to trastuzumab in drug-resistant cell lines. Mechanistic analyses indicated that SHP-1 might target STAT3 to inactivate p-STAT3 and reduce expression of its downstream signaling pathway. In conclusion, results presented here suggest that targeting SHP-1 may represent a potential approach to re-sensitize previously resistant cells to trastuzumab treatment.
Keywords
Breast cancer, Shp-1, HER-2, Trastuzumab, Stat3.
Introduction
Breast cancer (BC) is the most common malignant cancer threatening female health and longevity. Over the past decade, there has been an increase in breast cancer incidents, indicating that effective prevention and treatment methods are needed [1]. Different subtypes of breast cancer have been classified based on ER/PR and Her2 status, and each has its own unique recognizable phenotype and clinical outcome [2,3]. Amplification of human epidermal growth factor receptor 2 gene (HER-2) has been observed in 25%-30% of all BC cases [4].
HER-2-positive BC tumors often show the following characteristics: an aggressive disease, a poor prognosis, and a short overall survival (OS) [5,6]. Trastuzumab (Herceptin) was the first monoclonal antibody to be approved for targeted therapeutic use, either alone or in combination with the chemotherapy drug paclitaxel, in the treatment of metastatic HER-2-positive breast cancer [7]. Although many patients with metastatic HER-2+ breast cancer respond positively in clinical treatments to the first-line agent trastuzumab, virtually all patients eventually adapt to the anti-HER2 therapy and the disease progresses [8-10]. These observations indicate that resistance to trastuzumab in BC therapies is becoming more and more common.
SHP-1, an SH2 domain-containing cytosolic protein tyrosine phosphatase, is primarily expressed in hematopoietic stem cells functioning as a key regulator controlling intracellular phosphotyrosine levels in lymphocytes [11]. SHP-1 has been proposed as a candidate tumor suppressor gene for several cancers, such as lymphoma, breast cancer, and prostate cancer. Ordinarily, it plays a leading role in preventing oncogenic potentials and growth-promoting of tyrosine kinase. Thus, we raised the hypothesis that SHP-1 may reverse trastuzumab resistance via dephosphorylation.
In previous studies, we demonstrated that SHP-1 interacts with HER-2 and EGFR, functioning as a negative regulatory factor of tyrosine kinase activity through the inhibition of phosphorylation. Targeting HER-2, by combining SHP-1 overexpression and trastuzumab treatment, may improve response in HER-2-positive BC cells.
In this study, upregulation of SHP-1 expression was accomplished by transfecting with a lentivirus vector. We further explored the effect of SHP-1 on cell proliferation, cell migration, and overall improving sensitivity to trastuzumab. These studies determined whether SHP-1 can serve as a potential target for reversing resistance to trastuzumab. Furthermore, the molecular mechanism underlying the upregulated expression of SHP-1 in the resistant cells was addressed.
Materials and Methods
Cell lines and cell culture
The human mammary carcinoma cell lines SKBR3 was obtained from the Cell Bank of the Chinese Academy of Science (Xuhui, Shanghai, China). SKBR3 cells were maintained in the DMEM high glucose medium (Gibco, USA) supplemented with 10% fetal bovine serum (PAN, Germany). Trastuzumab-resistant cells (SKBR3-Pool2) were kindly provided by Dr. Qian Geng from Cancer Research Institute of Southern Medical University. SKBR3-Pool2 cells were maintained in a monolayer culture in DMEM/F12 (Dulbecco's modified Eagle's Medium, Gibco, USA) with 10% fetal bovine serum containing 70 μg/mL trastuzumab (Herceptin, Genentech, Inc) [12]. Both cell lines were incubated in a 37°C humidified atmosphere containing 95% air and 5% CO2 and split twice a week.
MTT assay
SKBR3 and SKBR3-Pool2 cells were cultured and counted and each test was conducted in triplicate. After mixing well, media/ cell solution was diluted to 5,000 cells per 200 μl. Cells were then plated onto a 96-well plate for 24 h. Cells were then grown in either control medium or the same medium containing trastuzumab for each treatment dosage (0 μg/ml control, 25, 50, 100, 200, 400, 800, and 1600 μg/ml) and incubated for another 96 h. Subsequently, 20 μl of MTT buffer reagent (5 mg/mL) was added into each well. After incubation for 4 h at 37°C, 100 μl of DMSO was added with gentle agitation for 10 min. After reading at 490 nm with a microplate reader, the percentages of surviving cells from each group relative to controls, defined as 100%, was determined by reduction of MTT.
Lentivirus transfection
Lentivirus vectors expressing GFP-(control) and GFP-SHP-1 were purchased from Biowit Inc. (Shenzhen, China). SKBR3- Pool2 cells were plated in each well of 24-well plates and allowed to adhere for 24 h. On the day of transfection, GFP- (control) or GFP-SHP-1 lentivirus-containing media (MOI=200) were thawed completely at room temperature.
Fresh medium containing polybrene (8 μg/mL) in a final volume of 1000 μl was added into the virus-containing media, which were used to replace the culture media of designated cells. After 24 h, virus-infected cells were selected by puromycin (2 μg/mL) for 72 h and subjected to required assays.
Colony formation assay
Briefly, cells were seeded in a 12-well plate with DMEM/F12 at 200 cells/well (Plate colony formation assays). Following 24 h of incubation at 37°C, drugs were added to the cells. After incubation of cells, the medium was replaced with the fresh complete medium in every 72 h. The incubation continued, allowing colonies to form for nearly 2 weeks until they were visible. Then, cells were fixed and stained with 2% crystal violet in ethanol for 15 min. The colony formation efficiency (CFE) was calculated by dividing the number of colonies by the number of cells plated and expressed as a percentage. Colony size and numbers were investigated using an inverted microscope and the results were analyzed by comparing the number of colonies of the control versus the treated cells.
Wound-healing assay
Cell migration is commonly evaluated by an in vitro scratch wound healing assay. The cells were seeded into 12-well plates in complete media containing 10% FBS to reach a confluency of 80%-90%. The nearly confluent monolayers of cells were manually scratched using a sterile micropipette tip, followed by washing with PBS to remove cellular debris. Then, new medium, containing designated drugs, was added to each well. The closure of the scratch was monitored under a phase contrast microscope 24 and 48 h after treatment. The percentage of inhibition was expressed using untreated wells at 100%.
Cell lysis and Western immunoblotting
Cells were trypsinized, washed twice with PBS, and lysed using a lysis buffer on ice for 30 min. Then cell lysates were cleared by centrifugation at 14,000 rpm for 15 min. Protein concentrations of the clear lysates was determined using the BCA Protein Determination Kit (Keygen Biotech, China). Equal quantities of each cell protein (30 μg) were separated on a 10%-12% SDS-PAGE gel and the proteins were transferred to a PVDF membrane (Millipore). The membrane was then blocked with 5% bovine serum albumin in TBS with Tween (TBST) and incubated with the appropriate primary antibodies overnight at 4°C. Membranes were incubated with secondary antibodies conjugated to horseradish peroxidase for 1 hour at room temperature. Bands were detected using enhanced chemiluminescence reagents according to the manufacturer’s instructions (Millipore, Inc.). For analysis of the Western blotting data, densitometric analysis was performed using Image J software, and band densities were normalized with respect to GAPDH (Cat. No. MB001, 1:5000, Bioworld) as the internal control. STAT3 (Cat. No. #9139, 1:1000, CST) and phosphorylated STAT3 (P-STAT3) mAbs (Cat. No. #9145, 1:2000, CST); c-Myc (Cat. No. ab32072, 1:5000, Abcam) and SHP-1 mAbs (Cat. No. ab2020, 1:800, Abcam); CyclinD1 polycolnal antibody (Cat. No. sc-718, 1:500, SANTA); MMP-9 mAbs (Cat. No. 10375-2-AP, 1:1000, Proteintech).
Statistical analysis
All experiments were repeated at least three times. Statistical analysis was performed using IBM SPSS statistical software (version 20.0). In each experiment, all samples were set up in triplicate, and the data were presented as the mean ± SD. Comparisons between 2 groups were made using a two-sided Student's t test. A value of P<0.05 is considered statistically significant.
Results
Identification of the phenotype of the trastuzumabresistant SKBR3 model
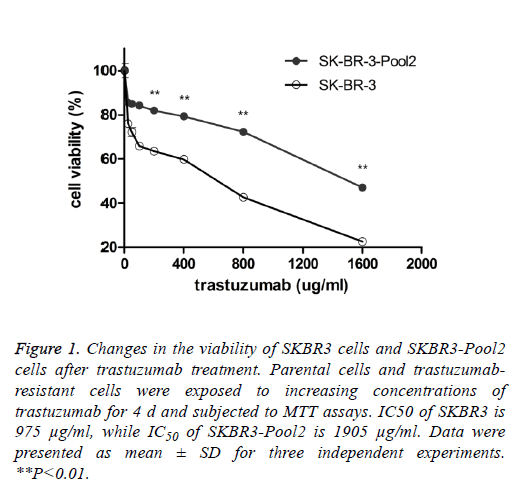
SKBR3 is a trastuzumab-sensitive breast cancer cell line while SKBR3-Pool2 is a trastuzumab-resistant cell line. SKBR3- Pool2 cell was maintained in the presence of 70 μg/mL trastuzumab. Cell proliferation (MTT) assays were performed and confirmed the resistant phenotype of the SKBR3-Pool2 cells. Consistent with previous studies characterizing trastuzumab-resistant breast cancer cells, growth inhibition of SKBR3-Pool2 cells was significantly lower than that of SKBR3 cells under the same trastuzumab medium conditions (Figure 1).
Figure 1: Changes in the viability of SKBR3 cells and SKBR3-Pool2 cells after trastuzumab treatment. Parental cells and trastuzumabresistant cells were exposed to increasing concentrations of trastuzumab for 4 d and subjected to MTT assays. IC50 of SKBR3 is 975 μg/ml, while IC50 of SKBR3-Pool2 is 1905 μg/ml. Data were presented as mean ± SD for three independent experiments. **P<0.01.
Stable SHP-1 overexpression promotes sensitivity to trastuzumab treament
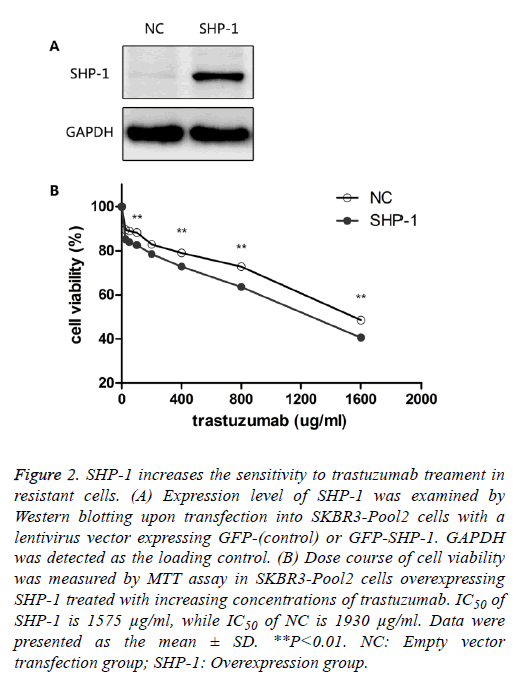
To identify the role of SHP-1 in breast cancer development, Yip et al. [13] investigated the expression of SHP-1 in human breast cancer cell lines and primary breast cancers. They found that the expression of SHP-1 mRNA correlated with ER status; high levels of this transcript were expressed in the ER-positive cell lines whereas the ER-negative lines expressed low levels. To further elucidate its biological role in HER-2 positive BC cells, SHP-1 vector and its control were respectively introduced into SKBR3-Pool2 cells. Efficiency of the SHP-1 overexpression was assessed by Western blotting. Significant increase in SHP-1 expression was observed in SKBR3-Pool2 cells treated with the SHP-1 vector compared with the control group (Figure 2A). Then, the sensitivity to trastuzumab of modified cells was examined by MTT assays. SKBR3 cell lines stably overexpressing SHP-1 exhibited significantly increased sensitivity to trastuzumab treatment. Cell viability 4 d after treatment at different concentrations of trastuzumab was calculated for cells with or without SHP-1 transfection. As show in Figure 2B, transfection with lentivirus containing SHP-1, but not the control vector, increased the levels of SHP-1 expression in SKBR3-Pool2 cells, leading to restoration of sensitivity to trastuzumab-mediated growth inhibition.
Figure 2: SHP-1 increases the sensitivity to trastuzumab treament in resistant cells. (A) Expression level of SHP-1 was examined by Western blotting upon transfection into SKBR3-Pool2 cells with a lentivirus vector expressing GFP-(control) or GFP-SHP-1. GAPDH was detected as the loading control. (B) Dose course of cell viability was measured by MTT assay in SKBR3-Pool2 cells overexpressing SHP-1 treated with increasing concentrations of trastuzumab. IC50 of SHP-1 is 1575 μg/ml, while IC50 of NC is 1930 μg/ml. Data were presented as the mean ± SD. **P<0.01. NC: Empty vector transfection group; SHP-1: Overexpression group.
SHP-1 inhibits cell migration and colony formation in SKBR3-Pool2 cells
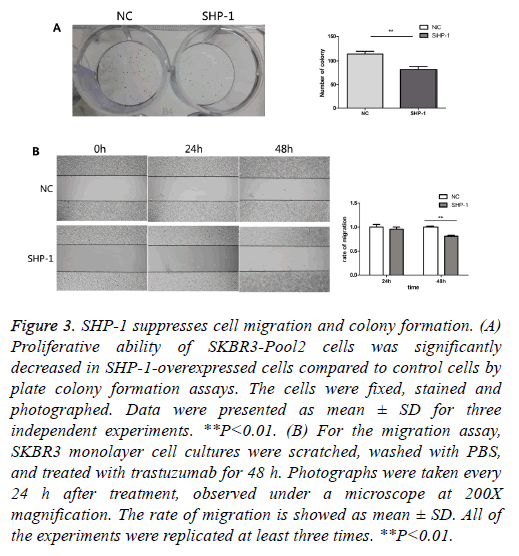
To further evaluate the biological effect of SHP-1, a colony formation assay was performed. Colony formation assays are a useful method for the evaluation of antitumor effects, thus this approach was used as a means to detect the function of SHP-1 in the colony formation of resistant cells. We compared the number of colonies in cells with overexpression of SHP-1 versus the control of SHP-1. As shown in Figure 3A, the colony formation number changed from 82 ± 10.583 in SKBR3-Pool2-SHP-1 cells to 114.67 ± 10.263 in parallel control cells after 2 weeks. Furthermore, it was investigated whether SHP-1 suppresses the migration of SKBR3-Pool2 cells by scratch wound healing assay. Obvious change in the rate of migration for 24 h was not observed in SHP-1-treated SKBR3-Pool2 cells. Nonetheless, the scarring area had less closure in SKBR3-Pool2-SHP-1 cells after 48 h, compared to transfected control cells (Figure 3B). As shown in Figure 3B, the rate of migration of SKBR3-Pool2-SHP-1 cells and parallel control cells was 0.81 ± 0.014 and 1 ± 0.015, respectively.
Figure 3: SHP-1 suppresses cell migration and colony formation. (A) Proliferative ability of SKBR3-Pool2 cells was significantly decreased in SHP-1-overexpressed cells compared to control cells by plate colony formation assays. The cells were fixed, stained and photographed. Data were presented as mean ± SD for three independent experiments. **P<0.01. (B) For the migration assay, SKBR3 monolayer cell cultures were scratched, washed with PBS, and treated with trastuzumab for 48 h. Photographs were taken every 24 h after treatment, observed under a microscope at 200X magnification. The rate of migration is showed as mean ± SD. All of the experiments were replicated at least three times. **P<0.01.
SHP-1 deregulates pSTAT3 in the STAT3 pathway to reverse trastuzumab-resistance and suppress cell proliferation and migration
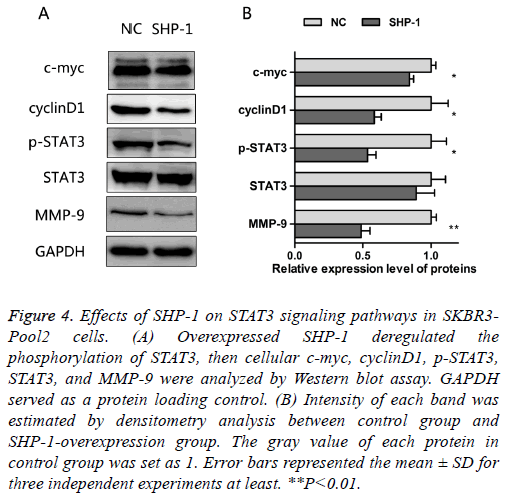
As noted above, SHP-1 not only increased the sensitivity to trastuzumab, but also suppressed the proliferation and migration in resistant cells. The oncogenic transcription factor, STAT3, is associated with the proliferation, metastasis and chemo-resistance of cancer cells [14]. Constitutive STAT3 activation is found in about 50~60% of breast tumors and associated with tumorigenesis and drug resistance [15-17]. In order to assess the effect of SHP-1 on this key element and some of its downstream targets, the expressions of STAT3, p- STAT3, c-myc, cyclinD1, and MMP-9 were examined by Western blotting. The results presented in Figure 4 show that SHP-1 decreased the levels of p-STAT3, c-myc, cyclinD1, and MMP-9; however, there was no significant change in the expression of STAT3, as compared with control modified cells. Moreover the levels of MMP-9, p-STAT3, and cyclinD1 were significantly reduced on SHP-1 overexpression, followed by the level of c-myc.
Figure 4: Effects of SHP-1 on STAT3 signaling pathways in SKBR3- Pool2 cells. (A) Overexpressed SHP-1 deregulated the phosphorylation of STAT3, then cellular c-myc, cyclinD1, p-STAT3, STAT3, and MMP-9 were analyzed by Western blot assay. GAPDH served as a protein loading control. (B) Intensity of each band was estimated by densitometry analysis between control group and SHP-1-overexpression group. The gray value of each protein in control group was set as 1. Error bars represented the mean ± SD for three independent experiments at least. **P<0.01.
Discussion
SHP-1 is a protein-tyrosine phosphatase (PTP) expressed in multiple cell lineages [18]. It is well documented that SHP-1 acts predominantly as a negative regulator in cell signal transduction pathways [19], which directly interacts with growth factors and other cytokine receptors to reduce phosphorylation of tyrosine residues in proteins [20]. Protein tyrosine kinases (PTKs) and protein tyrosine phosphatases (PTPs) keep the tyrosine phosphorylation level at a dynamic equilibrium. Any aberration in this balance can lead to a wide range of diseases because of the intracellular accumulation of tyrosine-phosphorylated proteins, which will cause abnormal cell proliferation and differentiation [21]. To study the specific function of SHP-1, lentivirus with SHP-1 was transfected into resistant cells. SKBR3-Pool2 cells expressing SHP-1 displayed a significant increase in SHP-1 expression compared with that of control cells expressing an empty vector. The biological role of SHP-1 in HER-2-positive BC-resistant cells was then investigated.
HER-2-positive BC presents with a unique epidemiology and molecular biology as well as clinical characteristics resulting in an aggressive disease, poor prognosis, and poorer response to chemotherapy. As a result, there is a low overall survival in patients [22]. HER-2 gene amplification has been identified in approximately 25-30% of BCs. Trastuzumab is the first monoclonal antibody to be approved for the treatment of HER2-overexpressing BC [23]. Even though trastuzumab combination chemotherapy holds promise for BC patients, the majority of patients who achieve an initial response to trastuzumab develop resistance within 1 year [12]. These observations indicate that acquired or secondary resistance to trastuzumab is also a major concern. Therefore, it is necessary to improve understanding of the molecular mechanisms that account for trastuzumab resistance. In the current study, the trastuzumab-resistance cell model has stable growth, and is suitable for further research investigating the role of SHP-1 in trastuzumab resistance. These studies demonstrated that resistant cells can withstand higher concentrations of trastuzumab than that of control cells by comparing the difference of drug sensitivity between sensitive cells and resistant cells under different concentrations of trastuzumab medium conditions.
SHP-1 is widely expressed in lymphomas, leukemias, and other tumors; however, the precise functions and targets of SHP-1 in BC cells are largely unknown. We have previously identified that SHP-1 suppressed cell proliferation, invasion, and metastasis in BC (unpublished data). Furthermore, we found that HER-2 is a potential substrate of SHP-1 based on co-association or heterodimerization. This study sought to further that work by emphasizing the role of SHP-1 in HER-2- positive and trastuzumab-resistant BC cells. The results indicated that overexpression of SHP-1 significantly restored the sensitivity to trastuzumab in resistant cells compared with the control group. SHP-1 was shown to reverse resistance and change drug sensitivity to trastuzumab Resistant cells typically showed characteristics of malignancy compared to sensitive cells, such as the potential for further proliferation or migration in the presence of chemotherapeutic drugs. In clinical applications of trastuzumab, patients with acquired resistance are more likely to have a tumor metastasis. This may reflect the enhanced ability of resistant cells to migrate under these conditions. Next, the role of SHP-1 in affecting the proliferation and migration of resistant cells under the trastuzumab exposure was explored. The results suggest that SHP-1 not only reduced the size and number of colonies formed but also suppressed the rate of migration in resistant cells. It may follow that as SHP-1 can inhibit the proliferation and migration of drug-resistant cells, it may also effectively control metastasis. This observation may provide more information regarding the role of trastuzumab for future treatment. Some studies about trastuzumab resistance, in the past few years, had been done in breast cancer. Many evidence suggests that the phosphorylation of STAT can be inhibited by overexpressing SHP-1. However, it is not clear about the mechenism of SHP-1 in the reversal of resistance to trastuzumab. The focus of our work is on the effect of SHP-1 in reversing trastuzumab resistance and the preliminary mechenism by which SHP-1 modulates BC cells sensitivity to trastuzumab.
The overexpression of SHP-1 increased in response to trastuzumab, and inhibited the proliferation and migration ability in resistant cells. However, little is known regarding the molecular mechanism of SHP-1 in the process of reversing resistance to trastuzumab. As a protein tyrosine phosphatase, SHP-1 takes part in regulation and control by phosphorylation and dephosphorylation, maintaining a state of equilibrium in the cell [24]. There are several signaling pathways that are closely related to the cellular process of phosphorylation or dephosphorylation in cancer cells. It is known that protein tyrosine phosphatases (PTPs) have been reported to negatively regulate STAT3 activation [25-27]. It has been documented that STAT3 hyperactivation was sufficient to confer trastuzumab resistance in HER-2-positive BC [28,29]. These studies suggest that pSTAT3 expression may be a critical point in signaling alteration promoting acquired trastuzumab resistance [28]. Collectively, the role of SHP-1 as a negative regulator led us to hypothesize that SHP-1 might decrease the expression of pSTAT3 via the STAT3 pathway. The results are consistent with our assumption demonstrating that SHP-1 participates in the reversal of trastuzumab resistance and metastasis through the suppression of pSTAT3 expression in our model. Decreased STAT3 phosphorylation suppresses the resistant cell’s progress by deregulating the expression of downstream target genes that control proliferation (cyclin D1 and c-myc) and migration (MMP-9).
In conclusion, these studies show that overexpression of SHP-1 can reverse drug resistance and reduce the ability of proliferation and migration in BC cells with trastuzumab resistance. In addition, we propose that SHP-1 inhibits the phosphorylation of STAT3 and deregulates its downstream signaling, suppressing trastuzumab resistance and metastasis in HER-2 positive BC. Initial targeted therapy combined with inhibition of STAT3 activation is a possible strategy to overcome trastuzumab resistance in the clinic. Targeting SHP-1 may represent a potential approach to re-sensitize resistant cells to trastuzumab treatment. The findings in this study reveal the beginning of the full understanding of the mechanisms of resistance or adaptation to trastuzumab. Further studies are required to disclose the control mechanism of SHP-1 in HER-2-positive BC and its potential as a therapeutic target in trastuzumab resistance.
Acknowledgment
We thank Dr. Rongcheng Luo and Dr. Rong Li for their helpful suggestions. We would like to thank Dr. Aiming Li and W.Y.F. for kind advice on conducting experiments and data analysis. We also acknowledge all the members of the Cancer Center for helpful discussions. This research work was supported in full by the grant from the National Natural Science Foundation of China (No.81272546 to Rong Li), Science and Technology Funding Projects of Guangdong (No. 2016ZC0056), Science and Technology Funding Projects of Guangzhou (No. 201604020009).
References
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66: 115-132.
- Network CGA. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61-70.
- Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res 2009; 7: 4-13.
- Ross JS, Slodkowska EA, Symmans WF, Pusztai L, Ravdin PM, Hortobagyi GN. The HER-2 receptor and breast cancer: ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist 2009; 14: 320-368.
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987; 235: 177-182.
- Witton CJ, Reeves JR, Going JJ, Cooke TG, Bartlett JM. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J Pathol 2003; 200: 290-297.
- Verma S, Joy AA, Rayson D, McLeod D, Brezden-Masley C, Boileau JF, Gelmon KA. HER story: the next chapter in HER-2-directed therapy for advanced breast cancer. Oncologist 2013; 18: 1153-1166.
- Nagata Y, Lan KH, Zhou X, Tan M, Esteva FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT, Hortobagyi GN, Hung MC, Yu D. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell 2004; 6: 117-127.
- Buzdar AU, Ibrahim NK, Francis D, Booser DJ, Thomas ES, Theriault RL, Pusztai L, Green MC, Arun BK, Giordano SH, Cristofanilli M, Frye DK, Smith TL, Hunt KK, Singletary SE, Sahin AA, Ewer MS, Buchholz TA, Berry D, Hortobagyi GN. Significantly higher pathologic complete remission rate after neoadjuvant therapy with trastuzumab, paclitaxel, and epirubicin chemotherapy: results of a randomized trial in human epidermal growth factor receptor 2-positive operable breast cancer. J Clin Oncol 2005; 23: 3676-3685.
- Chakrabarty A, Bhola NE, Sutton C, Ghosh R, Kuba MG, Dave B, Chang JC, Arteaga CL. Trastuzumab-resistant cells rely on a HER2-PI3K-FoxO-survivin axis and are sensitive to PI3K inhibitors. Cancer Res 2013; 73: 1190-200.
- Wu C, Sun M, Liu L, Zhou GW. The function of the protein tyrosine phosphatase SHP-1 in cancer. Gene 2003; 306: 1-12.
- Huang X, Gao L, Wang S, McManaman JL, Thor AD, Yang X, Esteva FJ, Liu B. Heterotrimerization of the growth factor receptors erbB2, erbB3, and insulin-like growth factor-i receptor in breast cancer cells resistant to herceptin. Cancer Res 2010; 70: 1204-1214.
- Yip SS, Crew AJ, Gee JM, Hui R, Blamey RW, Robertson JF, Nicholson RI, Sutherland RL, Daly RJ. Up-regulation of the protein tyrosine phosphatase SHP-1 in human breast cancer and correlation with GRB2 expression. Int J Cancer 2000; 88: 363-368.
- Abubaker K, Luwor RB, Escalona R, McNally O, Quinn MA, Thompson EW, Findlay JK, Ahmed N. Targeted disruption of the JAK2/STAT3 pathway in combination with systemic administration of paclitaxel inhibits the priming of ovarian cancer stem cells leading to a reduced tumor burden. Front Oncol 2014; 4: 75.
- Armanious H, Gelebart P, Mackey J, Ma Y, Lai R. STAT3 upregulates the protein expression and transcriptional activity of beta-catenin in breast cancer. Int J Clin Exp Pathol 2010; 3: 654-664.
- Hart JR, Liao L, Yates JR, Vogt PK. Essential role of Stat3 in PI3K-induced oncogenic transformation. Proc Natl Acad Sci U S A 2011; 108: 13247-13252.
- Chung SS, Giehl N, Wu Y, Vadgama JV. STAT3 activation in HER2-overexpressing breast cancer promotes epithelial-mesenchymal transition and cancer stem cell traits. Int J Oncol 2014; 44: 403-411.
- Yi TL, Cleveland JL, Ihle JN. Protein tyrosine phosphatase containing SH2 domains: characterization, preferential expression in hematopoietic cells, and localization to human chromosome 12p12-p13. Mol Cell Biol 1992; 12: 836-846.
- Tsui FW, Martin A, Wang J, Tsui HW. Investigations into the regulation and function of the SH2 domain-containing protein-tyrosine phosphatase, SHP-1. Immunol Res 2006; 35: 127-136.
- Wu C, Guan Q, Wang Y, Zhao ZJ, Zhou GW. SHP-1 suppresses cancer cell growth by promoting degradation of JAK kinases. J Cell Biochem 2003; 90: 1026-1037.
- Siminovitch KA, Lamhonwah AM, Somani AK, Cardiff R, Mills GB. Involvement of the SHP-1 tyrosine phosphatase in regulating B lymphocyte antigen receptor signaling, proliferation and transformation. Curr Top Microbiol Immunol 1999; 246: 291-297.
- Paik S, Bryant J, Tan-Chiu E, Yothers G, Park C, Wickerham DL, Wolmark N. HER2 and choice of adjuvant chemotherapy for invasive breast cancer: National Surgical Adjuvant Breast and Bowel Project Protocol B-15. J Natl Cancer Inst 2000; 92: 1991-1998.
- Carter P, Presta L, Gorman CM, Ridgway JB, Henner D, Wong WL, Rowland AM, Kotts C, Carver ME, Shepard HM. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc Natl Acad Sci U S A 1992; 89: 4285-4289.
- Lopez-Ruiz P, Rodriguez-Ubreva J, Cariaga AE, Cortes MA, Colas B. SHP-1 in cell-cycle regulation. Anticancer Agents Med Chem 2011; 11: 89-98.
- Chun J, Li RJ, Cheng MS, Kim YS. Alantolactone selectively suppresses STAT3 activation and exhibits potent anticancer activity in MDA-MB-231 cells. Cancer Lett 2015; 357: 393-403.
- Yu ZY, Huang R, Xiao H, Sun WF, Shan YJ, Wang B, Zhao TT, Dong B, Zhao ZH, Liu XL, Wang SQ, Yang RF, Luo QL, Cong YW. Fluacrypyrim, a novel STAT3 activation inhibitor, induces cell cycle arrest and apoptosis in cancer cells harboring constitutively-active STAT3. Int J Cancer 2010; 127: 1259-1270.
- Han Y, Amin HM, Franko B, Frantz C, Shi X, Lai R. Loss of SHP1 enhances JAK3/STAT3 signaling and decreases proteosome degradation of JAK3 and NPM-ALK in ALK+ anaplastic large-cell lymphoma. Blood 2006; 108: 2796-2803.
- Li G, Zhao L, Li W, Fan K, Qian W, Hou S, Wang H, Dai J, Wei H, Guo Y. Feedback activation of STAT3 mediates trastuzumab resistance via upregulation of MUC1 and MUC4 expression. Oncotarget 2014; 5: 8317-8329.
- Sonnenblick A, Brohee S, Fumagalli D, Vincent D, Venet D, Ignatiadis M, Salgado R, Van den Eynden G, Rothe F, Desmedt C, Neven P, Loibl S, Denkert C, Joensuu H, Loi S, Sirtaine N, Kellokumpu-Lehtinen PL, Piccart M, Sotiriou C. Constitutive phosphorylated STAT3-associated gene signature is predictive for trastuzumab resistance in primary HER2-positive breast cancer. BMC Med 2015; 13: 177.