ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2015) Volume 26, Issue 2
Screening methods using clopidogrel by genotyping CYP2C19 cytochrome and ABCB1 gene in patients with acute coronary syndromes.
1Faculty of Medicine, University of Medicine and Pharmacy Grigore T.Popa Iasi, 16 Universității St., Iasi, 700115, Romania
2CI Negoiţă 1st Cardiology Department, Sf. Spiridon Emergency Districtual Clinical Hospital, 1 Piața Independenței St., Iasi,700111, Romania
3Faculty of Pharmacy, University of Medicine and Pharmacy Grigore T.Popa Iasi, 16 Universității St., Iasi, 700115, Romania
4Immunology Department-Molecular Medicine Platform; Regional Oncology Institute, Molecular Biology Laboratory, 3 Vasile Conta St., Iasi, 700111, Romania
5Arcadia Hospital, 2 Cicoarei St., Iasi, 700620, Romania
- *Corresponding Author:
- Ana Clara Aprotosoaie
University of Medicine and Pharmacy
Grigore T.Popa Iasi Universităţii St. No16
Iasi Romania
Accepted date: January 18 2015
Clopidogrel resistance is a phenomenon which has clinical implications related to the evolution and unfavorable prognosis of patients with acute coronary syndromes (ACS). Although different mechanisms for Clopidogrel resistance have been suggested, genetic polymorphism of liver enzyme CYP2C19 is on of the most prominent. The aim of the study was to determine variations in response to Clopidogrel in a group of 96 patients diagnosed with acute coronary syndromes (ACS), by genotyping the CYP2C19 cytochrome and the ABCB1 gene. CYP2C19 and ABCB1 polymorphisms were investigated using two methods: TaqMan Assay and High Resolution Melting (HRM) technique. Depending on the genotypic constellation of the alleles studied for CYP2C19 gene, patients were divided into several prediction categories: normal metabolizer (6.25%); intermediate metabolizer (0%); slow metabolizer (17.70%); very fast metabolizer (60.41%) and unpredictable metabolizer (15.62%). HRM technique may be a viable alternative to determine the CYP2C19*2, CYP2C19*17 and ABCB1 polymorphisms, being less expensive than TaqMan Assay.
Keywords
Clopidogrel, resistance, genetic polymorphisms, TaqMan, HRM
Introduction
Resistance to Clopidogrel, a concept that dates back several years, has two implications: a clinical one – related to the evolution and unfavorable prognosis of patients with acute coronary syndromes (ACS) and the second one-practical, that enforces introduction on the market of new antiplatelet agents lacking the resistance phenomenon [1-5]. One of the key enzymes involved in Clopidogrel metabolism, CYP2C19 cytochrome, has many isoforms that determine variations in the biotransformation of the prodrug into active metabolites [6-10]. Depending on the conversion rate of Clopidogrel, patients may be slow or fast metabolizers: some present hipo-response regarding the blocking of platelet clotting (followed by increased risk of thrombosis) and others present hiper-response (increased risk of bleeding) [11-15]. The so-called "genetic resistance" to clopidogrel is followed by a dramatic increase in the incidence of intrastent thrombosis in those patients as well as the recurrence of the subsequent ischemic events [2,6,16]. Besides the platelet function tests, genetic assays regarding the P450 cytochrome and its isoforms are the major methods for detecting resistance to clopidogrel [12,13]. This study aimed to determine variations in response to Clopidogrel in a group of patients diagnosed with ACS, by genotyping the CYP2C19 cytochrome and the ABCB1 gene using TaqMan Assay and High Resolution Melting (HRM) technique.
Materials and methods
The study included a group of 96 patients (15 women and 81 men, aged between 45-85 years) admitted to the Cardiology Clinic of the St. Spiridon Emergency Districtual Clinical Hospital Iasi during the period January - December 2012. The patients eligible for the study met the following criteria: a) clinical – previous anginal chest pain present during hospitalization or a history of severe myocardial infarction or previously diagnosed coronary graphic changes with or without subsequent angioplasty; b) paraclinical - suggestive electrocardiographical changes - subendocardic or subepicardic ischemic lession or recent branch block associated with changes in myocardic cytolysis enzymes and positive troponin. All patients in the study received platelet antiaggregant therapy with Clopidogrel, associated sometimes with aspirin. Paraclinical investigations included: analysis of associated risk factors (glycemia, uric acid, complete lipid profile), evaluation of renal function and electrolytes, electrocardiogram 12 derivations, echocardiogram (2D, Mmod and Doppler), complete blood count and coagulation tests. Patients were initially evaluated (at the inclusion in the study) following the complete check-up mentioned above. All patients were previously informed about the purpose of the study, those who agreed to be included in the study signed an informed consent and then a blood sample was taken for DNA extraction.
DNA isolation
The extractions were carried out by using Wizard Genomic DNA Purification - Promega Kit (Promega Inc., Madison, WI, USA). The DNA was quantified and then aliquoted and labeled in order to achieve DNA collection in Molecular Biology Laboratory (Regional Institute of Oncology, Iasi). Some of the DNA was stored at -20°C for the PCR experiments. The other samples were frozen at -80°C.
Genotype analysis
TaqMan assay (Applied Biosystems, Life Technologies, Pleasanton, CA, USA), used for the aimed single nucleotide polymorphism (SNP), is based on the allelic discrimination by Real-Time PCR in a TaqMan system. The same set of primers and one specific fluorescence probe are used for each allele. Through the degradation of the probes by the polymerase (with 5'-3' exonuclease properties) during amplification, a specific fluorescence will be released (only by the hybridized probe). This allows allelic discrimination as a color code. The probes used for the wild allele had a Reporter-VIC fluorescence, and the probes for the mutant allele had a Reporter-FAM fluorescence. All the probes that were used had a MGB unit (Minor Grove Binder) to the 3' end. TaqMan probes that have MGB incorporated at the 3' end have a very high specificity for the specific allele, therefore they are ideal for allelic discrimination experiments. MGB molecules are caught on the minor notch of the double helix, improving the hybridization process and stabilizing the probe matrix complex.
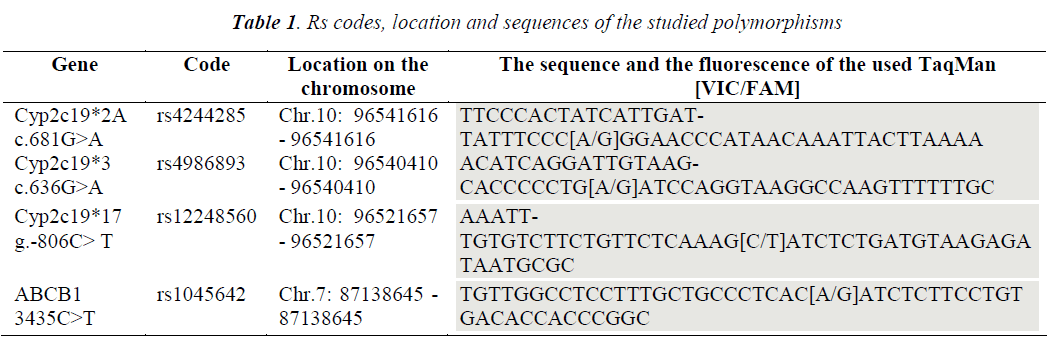
The current study used specific probes for mutant alleles *2A, *3, *17 of the CYP2C19 gene versus specific probes for the wild allele, as well as probes for mutant allele 3435C>T versus wild allele of the ABCB1 gene (Table 1). The amplification was performed using the Real Time PCR MX3005P Stratagene/Agilent thermal cycler. The amplification program (40 cycles): 95°C-10 min; 92°C-15 sec; 60º C – 1 min - acquisition on the FAM and VIC channels throughout the elongation. Simultaneously with the optimization and the determinations made through the TaqMan Assay, it was attempted to identify an alternative method with lower costs, that could make allelic discrimination with a confidence of over 90%, and it was decided to approach the High Resolution Melting technique (HRM). HRM is a technique which analyzes the melting curves of the fragments of interest post-PCR amplification (amplicons) (Fig. 1). The method is based on using the dissociation melting curves of the products of interest, being available due to the improvement of the dyes that emit fluorescence while binding on doublestranded DNA and the evolution of RealTime PCR equipment and the analysis programs associated to each machine. Discrimination between the types of polymorphisms by the dissociation curve is based on the composition of the amplicon, its length, the content of CG and the complementarity of the two strands. HRM begins with a PCR amplification of the region of interest, in the presence of dyes that bind to the double-helix and emit fluorescence.
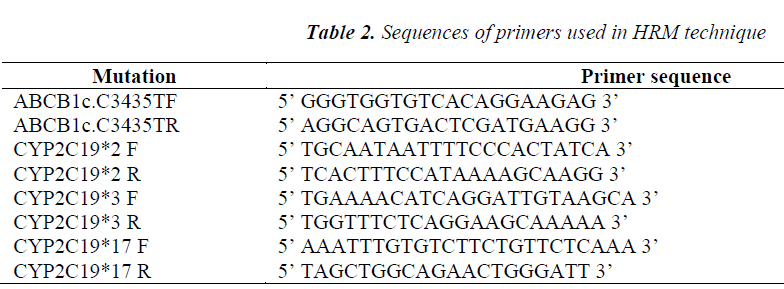
These fluorochromes emit a very high fluorescence when bound to double-stranded DNA (dsDNA) and they emit a very weak fluorescence when they are not bound to dsDNA. The amplification is followed then by a melting curve at a very high resolution, made of very narrow steps of temperature (0.1ºC) and of the reading of a very large number of fluorescent events for each step with a very high precision. When dsDNA is dissociated into component strands, the fluorochrome is released leading to a decrease in the overall fluorescence from the tube. The result is obtaining a melting curve characteristic for each amplicon. The HRM reaction can take place in a single step (PCR + MELT) or the PCR reaction can be performed in a standard thermal-cycler, and only HRM in a Real-Time PCR. Genotyping through HRM was performed for all ABCB1c polymorphisms: C3435T (rs1045642), CYP2C19*2 (c.G681A; rs4244285), CYP2C19*3 (c.G636A, rs4986893), and CYP2C19*17 (c.C806T, rs12248560), using RealTimePCR RotorGene 6000® (Qiagen, Courtaboeuf, France) and primers from Table 2.
The design of primer sequences was performed using„ Primer 3 imput” software. PCR reaction was done with addition of Bryte Green (Promega) fluorochrome. The conditions of PCR amplification were enzyme activation at 95ºC – 10 min followed by 40 cycles of: denaturation 95ºC - 15 sec, primers hybridization (different temperature depending on the melting temperature of each primer set)-15 sec, elongation 72ºC - 40 sec. The reading of fluorescence was made after every cycle of amplification on the Green channel. The curve of dissociation was achieved at a ramp from 70 to 94°C. The confidence threshold was established at 90%. HRM curves were normalized using pre-and post melting regions.
Results
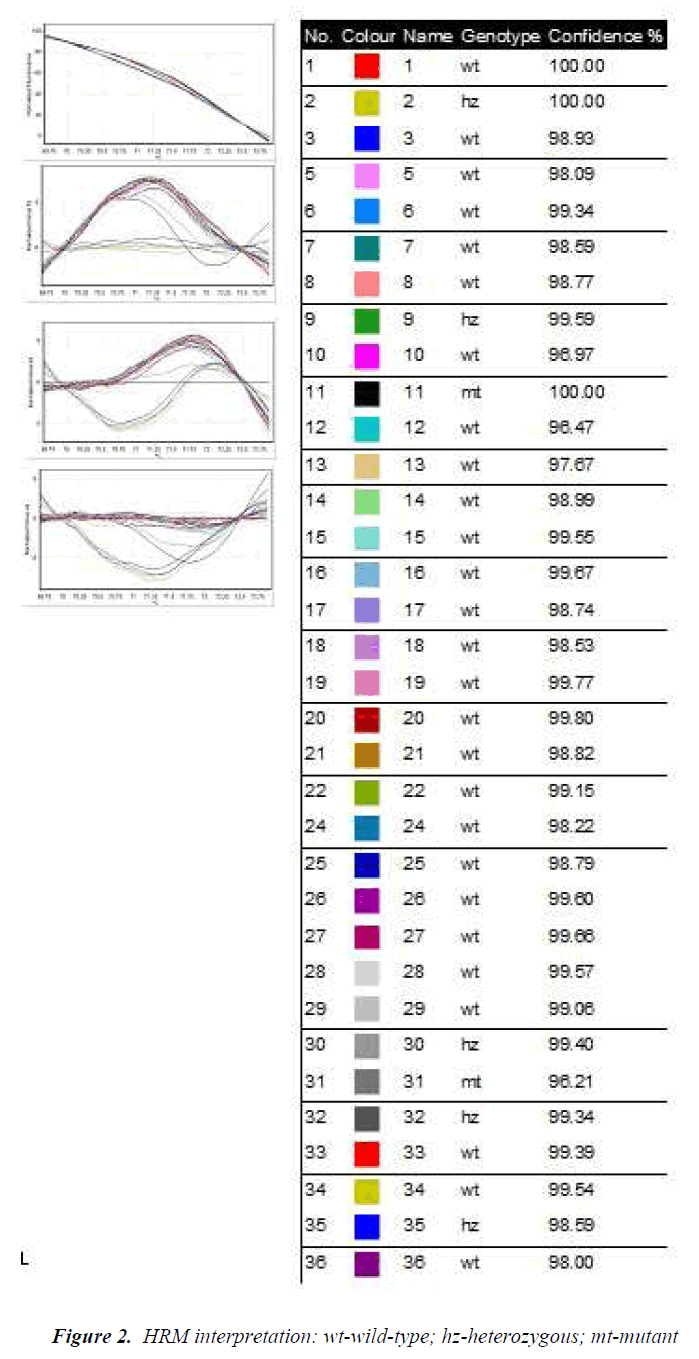
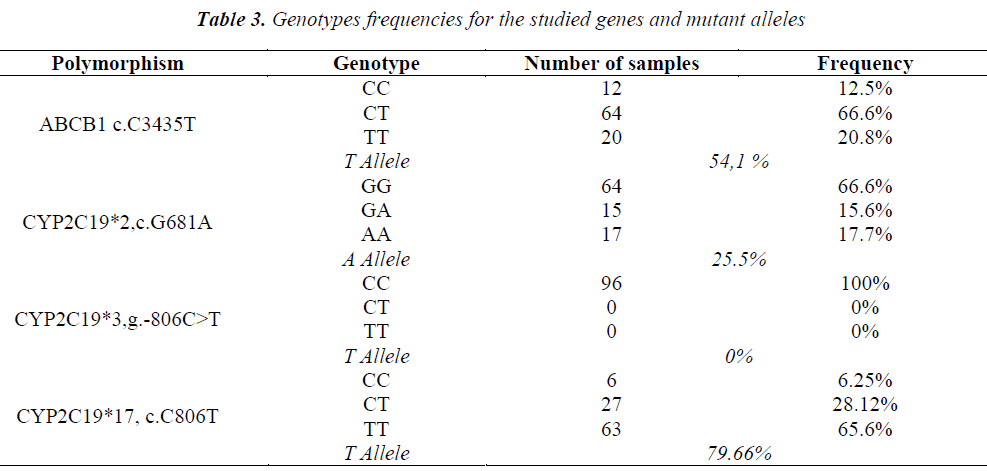
The genotypes obtained by TaqMan assay are shown in Table 3. Also, the amplification by BrytGreen master-mix was performed for the HRM technique for the 96 patients. Very good results were obtained from the first stage of the study for CYP2C19*2 and CYP2C19*17 polymorphisms for a group of 36 patients (Fig. 2). Primers functioned for these polymorphisms, giving specific amplicons (without non-specific binding and dimerization), and the results were 100% correlated with the genotyping through TaqMan. The obtained allelic discrimination confidence was of over 95%.
100% correlation between TaqMan genotyping and the one achieved through HRM was also confirmed by continuing RealTime PCR amplifications for the rest of the lot as well (44 patients). For the CYP2C19*3 and ABCB1 allele, the primers set initially were not suitable, forming nonspecific amplifications, that in a nonspecific RealTimePCR (with fluorochromes emitting fluorescence by conjugation with dsDNA) can alter the results.The design was made for other two sets of primers (through the primer 3 imput program). With these primer sets, optimal amplification we achieved. For the ABCB1 gene, where samples of wild-type (wt), heterozygous (hz) and mutant (mt) were previously determined (by TaqMan), the parameters of analysis were correctly set. Thus, the confirmation of TaqMan genotyping (100% correlation) was successful for all 96 patients. For the CYP2C19*3 allele, the RealTime amplification has been optimized so that it does not result in nonspecific binding (the appearance of dimers), but because there was no evidence of hz and mt control, parameter settings for the HRM analysis could not be performed.
Discussion
CYP2C19 cytochrome contributes significantly to the metabolizing of clopidogrel [17] and it influences the therapeutic response. At least 25 polymorphisms have been described for the gene encoding CYP2C19, of which the CYP2C19*2 allele (exon 5) is the best known and studied. It appears that it induces a significant drop in the concentration of the serum active metabolite and thus it reduces the inhibition of platelet aggregation [5]. Patients who have the CYP2C19*2A allele and who are treated with clopidogrel seem to have an increased risk for recurent ischemic events [11]. In addition, the presence of the CYP2C19*3A allele, in conjunction with the CYP2C19*2A allele, results in an increased risk of intrastent acute thrombosis [18]. On the contrary, the CYP2C19*17 allele was associated with an exacerbation of the enzymatic activity, being the allele with a ”gain of function”, that can increase the risk of bleeding during treatment with clopidogrel [15]. Polymorphisms that induce loss of function (CYP2C19*2, *3) were investigated in the present study (the most frequent in European statistics), as well as those that potentiate it (CYP2C19*17). Since the C3435T (rs1045642) allelic version of the ABCB1 gene influences the intestinal absorption of clopidogrel in patients with cardiovascular diseases [19], this polymorphism was also investigated. The genotyping results obtained by both TaqMan and HRM assays were similar. Depending on the genotypic constellation of the alleles investigated for CYP2C19 gene, patients were divided into several prediction categories: 1) normal metabolizer (N): wild-type CYP2C19*2*3*17; 2) slow metabolizer (MS): *2 /*2, *2/*3, *3/*3, indifferent of also being heterozygous or homozygous on *17; 3) very fast metabolizer (MFR): homozygous or heterozygous for *17 and wild-type for *2 and *3; 4) unpredictable metabolizer (MN): heterozygous or homozygous on *17 and heterozygous on *2 and/or *3. Regarding the corroboration of the genotyping results with the serological analyzes and the clinical picture, in the investigated group there were identified 6.25% patients with normal metabolizing phenotype, 17.70% slow metabolizing patients, 60.41% patients with very fast metabolizing phenotype and 15.62% patients with unpredictable phenotype. No patient was identified with an intermediate phenotype.
The proportion of normal metabolizer patients (wild-type CYP2C19*2*3*17) in this study is much lower compared to the specialized literature, which stresses the need for such studies on much larger groups of patients. The absence of the *3 allele in our study does not exclude the existence of individuals with slow metabolizing or intermediate phenotype (heterozygous for *2 and/or *3 and wild-type *17) resulting from the presence of other alleles with loss of function. Being relatively less expensive than TaqMan Assay, HRM technique would allow an easier access for a wider number of patients with acute coronary syndromes, that are treated with Clopidogrel.
Acknowledgements
Scientific research sponsored by University of Medicine and Pharmacy “Grigore T. Popa” of Iasi, based on Contract nr. 28214/2011.
References
- Angiolillo DJ, Fernandez-Ortiz A, Bernardo E, et al. Variability in individual responsiveness to clopidogrel: clinical implications, management, and future perspectives. J Am Coll Cardiol 2007; 49: 1505-1516.
- Eisenstein EL, Anstrom KJ, Kong DF, et al. Clopidogrel use and long-term clinical outcomes after drug-eluting stent implantation. JAMA 2007; 297: 159-168.
- Husted S. Evidence-based prescribing and adherence to antiplatelet therapy-how much difference do they make to patients with atherothrombosis? Int J Cardiol 2009; 134: 150-159.
- Nguyen TA, Diodati JG, Pharand C. Resistance to clopidogrel: a review of the evidence. J Am Coll Cardiol 2005; 45: 1157-1164.
- Simon T, Verstuyft C, Mary-Krause M, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med 2009; 360: 363-375.
- Bonello L, Bonello-Palot N, Armero S, et al. Impact of P2Y12-ADP receptor polymorphism on the efficacy of clopidogrel dose-adjustment according to platelet reactivity monitoring in coronary artery disease patients. Thromb Res 2010; 125: 167-170.
- Frere C, Cuisset T, Morange PE, et al. Effect of cytochrome P450 polymorphisms on platelet reactivity after treatment with clopidogrel in acute coronary syndrome. Am J Cardiol 2008; 101: 1088-1093.
- Giusti B, Gori AM, Marcucci R, et al. Relation of cytochrome P450 2C19 loss-of-function polymorphism to occurrence of drug-eluting coronary stent thrombosis. Am J Cardiol 2009; 103: 806-811.
- Paveliu MS, Bengea S, Paveliu FS. Individualized drug response related to genetic variations of cytochrome P450 isoforms and other enzymes. Farmacia 2010; 58: 245-254.
- Sibbing D, Stegherr J, Latz W, et al. Cytochrome P450 2C19 loss-of-function polymorphism and stent thrombosis following percutaneous coronary intervention. Eur Heart J 2009; 30: 916-922.
- Bonello L, Palot-Bonello N, Armero S, et al. Impact of loading dose adjustment on platelet reactivity in homozygotes of the 2C19 2* loss of function polymorphism. Int J Cardiol 2010; 145: 165-166.
- Costache II, Rusu C, Ivanov I, et al. Clopidogrel resistance-risk factor in patients with acute coronary syndromes. Rev.Med.Chir.Soc.Med.Nat. Iaşi 2012; 116: 383-389.
- Costache II, Rusu C, Ivanov I, et al. Impact of clopidogrel response on the clinical evolution in patients with acute coronary syndromes. Rev Med-Chir Soc Med Nat Iaşi 2012; 116: 962-968.
- Mega JL, Close SL, Wiviott SD, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med 2009; 360: 354-362.
- Sibbing D, Koch W, Gebhard D, et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation 2010; 121: 512-518
- Gurbel PA, Bliden KP, Hiatt BL, et al. Clopidogrel for coronary stenting: response variability, drug resistance, and the effect of pretreatment platelet reactivity. Circulation 2003; 107: 2908-2913.
- Kazui M, Nishiya Y, Ishizuka T, et al. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos 2010; 38: 92–99.
- Harmsze AM, van Werkum JW, Ten Berg JM, et al. CYP2C19*2 and CYP2C9*3 alleles are associated with stent thrombosis: a case-control study. Eur Heart J 2010; 31: 3046-3053.
- Taubert D, von Beckerath N, Grimberg G, et al. Impact of P-glycoprotein on clopidogrel absorption. Clin Pharmacol Ther 2006; 80: 486-501.