ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 9
Regulatory mechanism of extracellular signal-regulated kinase signal pathway for DNA methylation in lung carcinoma cells
Yu Sun1, Wen-Zhou Liu1, Tao Liu1, Xu Feng2, Nuo Yang2 and Hua-Fu Zhou2*
1Department of Cardio-Thoracic Surgery, the Second Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, PR China
2Department of Cardio-Thoracic Surgery, the First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi, PR China
- *Corresponding Author:
- Hua-Fu Zhou
Department of Cardio-Thoracic Surgery
The First Affiliated Hospital of Guangxi Medical University
PR China
Accepted date: December 11, 2017
DOI: 10.4066/biomedicalresearch.29-17-3471
Visit for more related articles at Biomedical ResearchObjective: This study aimed to discuss the regulatory mechanism of Extracellular Signal-Regulated Kinase (ERK) signal pathway for DNA methylation in lung carcinoma cells.
Methodology: Human lung carcinoma cells were cultured, and the cells in logarithmic phase were inoculated in 96-hole plates and divided into three groups A-C. Six complex holes were present in each group. Phosphoric buffer salt solution, agonist epidermal growth factor, and blocker U1026 solution of ERK pathway were added into groups A-C, with a total amount of 25 μmol/L. Then, they were continuously treated for 48 h. Methyl thiazol tetrazolium method was used to determine cell proliferation rate. Fluorescent quantitative Polymerase Chain Reaction (qPCR) method was used to test the expression quantities of DNA transmethylase 1 (Dnmt1), DNA transmethylase 3a (Dnmt3a), DNA transmethylase 3b (Dnmt3b), tumor suppressor gene (p16), and Ras association domain family gene (RASSF1A) mRNA.
Results: Significant differences exist among groups A-C in cell proliferation rate, as well as in Dnmt1, Dnmt3a, Dnmt3b, p16, and RASSF1A mRNA, and their protein expression levels (P<0.05). Cell proliferation rate and the expression quantities of Dnmt1, Dnmt3a, and Dnmt3b mRNA and their protein expression levels in group B were significantly higher than those in groups A and C (P<0.05). By contrast, the expression quantities of p16 and RASSF1A mRNA and their protein expression levels were significantly lower than those in groups A and C (P<0.05). Cell proliferation rate and the expression quantities of Dnmt1, Dnmt3a, and Dnmt3bmRNA and their protein expression levels in group A were significantly higher than those in group C (P<0.05).
Conclusion: ERK signal pathway blocked both proliferation rate of lung carcinoma cells and DNA methylation level because of the down-regulation of Dnmts mRNA and their protein expression levels and the up-regulation of cancer suppressor genes p16 and RASSF1A mRNA and their protein expression levels.
Keywords
Lung carcinoma, Extracellular regulation of protein kinase signal pathway, DNA methylation
Introduction
Lung carcinoma, the most common malignant tumor, has become a leading cause for human death with morbidity still being on the rise [1]. Epigenetics has exerted regulatory effect on the genesis and development of various cancers, including DNA methylation modification, histone modification, chromosome modification, and non-coding RNA [2,3]. DNA methylation modification plays a significant role in the genesis of lung carcinoma. DNA methylation and extracellular regulated protein kinase-motogen-activated protein kinases signal transduction pathway mediate or regulate relevant genetic expressions in the genesis and development of lung carcinoma [4]. However, further study is needed on the regulatory mechanism of ERK signal pathway for the DNA methylation of lung carcinoma cells [5]. In the current study, this was carried out by applying ERK signal pathway agonist and blocker in cells in vitro.
Materials and Methodology
Materials
Human carcinoma cell stain A549 (Shanghai Institute of Biochemistry and Cell Biology, CAS); RMPI 1640, DMEM culture medium, fetal calf serum, and Trizol total RNA extraction kits (Life Technologies, USA); agonist epidermal growth factor (EGF) and blocker U1026 of ERK pathway (Sigma, USA); real-time fluorescent quantitative polymerase chain reaction (qPCR) and reverse transcription kits (Takara, China); cell lysis buffer IP, Dnmt1, Dnmt3a, and Dnmt3b mice anti-human McAb (monoclonal antibodies) (Santa Cruz, USA); and horse radish peroxidase and goat anti-rabbit polyclonal antibodies (Chembiochem, USA) were used for the experiments.
Methodology
Detection of cell proliferation rate through methyl thiazol tetrazolium (MTT) method: After 48 h, 5 mg/mL 20 μL MTT solution was added into each hole for 4 h of continuous culturing. After the culture solution was abandoned, 150 μL dimethylsulfoxide was added into each hole for 10 min of decolorization and shaking reaction. Enzyme-linked immunosorbent assay was used to detect the absorbance (optical density value) at 570 nm. Holes containing only the culture medium solution constituted the control group, which was used to detect cell proliferation rate.
Detection of mRNA expression through real-time fluorescent qPCR: After 48 h, Trizol method was used to extract cellular total RNA. Agarose electrophoresis and ultraviolet (UV) spectrophotometer were used to determine RNA purity and concentration, respectively. RNA at 10 μg was reversely transcribed into cRNA according to the instructions of the reverse transcription kit and put under reaction under real-time fluorescent qPCR. The primers of Dnmt1, Dnmt3a, Dnmt3b, p16, and RASSF1A were designed and synthesized by the Shanghai Bioengineering Co., Ltd.
Detection of methylation level through methylation-specific PCR: After 48 h, trypsinization occurred in cells in the three groups, and these cells were collected into 1.5 ml centrifugal tube for 5 min of centrifugation at a speed of 13,000. The supernatant was discarded, and follow-up operation was implemented according to the procedure shown in the instructions of the DNA extraction kit. Agarose electrophoresis and UV spectrophotometer were used to detect DNA purity and concentration, respectively. The promoter sequences of tumor suppressor genes p16 and RASSF1A were searched according to UCSU database. Methylated CpG islands in p16 and RASSF1A promoter regions were predicted. Methylationspecific primers and non-methylated primers were then designed.
Statistical analysis
SPSS17.0 software was used for analysis, and data were expressed by mean ± standard deviation (SD). One-way ANOVA and Dunnett t-test were used for analysis. P<0.05 meant significant difference.
Results
Cell proliferation rate
Comparative differences among groups A-C in cell proliferation rate were significant (P<0.05), where cell proliferation rate in group B was significantly higher than those in groups A and C (P<0.05), but that in group A was significantly higher than that in group C (P<0.05).
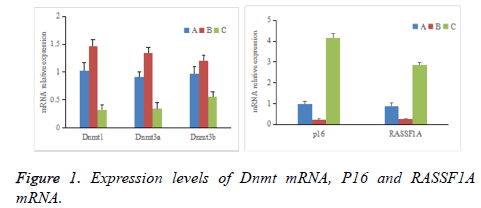
Expressions of Dnmt1, Dnmt3a, Dnmt3b, p16, and RASSF1A mRNA
As shown in Figure 1, comparative differences among groups A-C in the expression levels of Dnmt1, Dnmt3a, Dnmt3b, p16, and RASSF1A mRNA were significant (P<0.05). The expression levels of Dnmt1, Dnmt3a, and Dnmt3b mRNA in group B were significantly higher than those in groups A and C (P<0.05), whereas the expression levels of p16 and RASSF1A mRNA were significantly lower than those in groups A and C (P<0.05). The expression levels of Dnmt1, Dnmt3a, and Dnmt3b mRNA in group A were significantly higher than those in group C (P<0.05), whereas the expression levels of p16 and RASSF1A mRNA were significantly lower than those in group C (P<0.05).
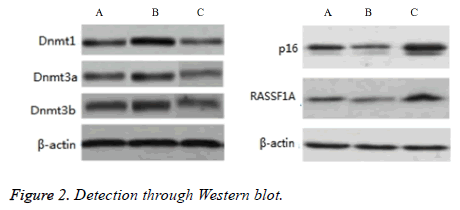
Protein expression levels of Dnmt1, Dnmt3a, Dnmt3b, p16, and RASSF1A
Comparative differences among groups A-C in the protein expression levels of Dnmt1, Dnmt3a, Dnmt3b, p16, and RASSF1A were significant (P<0.05). The protein expression levels of Dnmt1, Dnmt3a, and Dnmt3b in group B were significantly higher than those in groups A and C (P<0.05), but the protein expression levels of p16 and RASSF1A were significantly lower than those in groups A and C (P<0.05). The protein expression levels of Dnmt1, Dnmt3a, and Dnmt3b in group A were significantly higher than those in group C (P<0.05), but the protein expression levels of p16 and RASSF1A were significantly lower than those in group C (P<0.05), as shown in Figure 2.
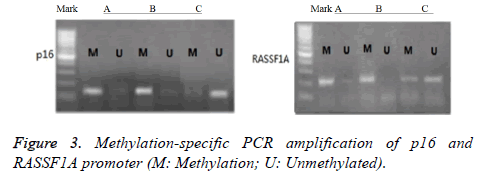
Methylation analysis of p16 and RASSF1A promoters
As shown in Figure 3, the detection showed that the promoter regions of tumor suppressor genes p16 and RASSF1A in groups A and B presented methylation status, and the amplified bands of methylated primers in group B were more apparent. The p16 promoter regions in group C presented un-methylated status. Some RASSF1A promoter regions presented methylation status, whereas others presented demethylation status.
Discussion
Through the detection of cell proliferation rating among the three groups, we found the following results. Cell proliferation rate in group B was the highest, but that in group C was the lowest. Blocking ERK pathway could inhibit the proliferation of lung carcinoma cells. Based on a study on the expression levels of Dnmt1, Dnmt3a, Dnmt3b, p16, and RASSF1A mRNA through fluorescent qPCR, the expression levels of Dnmt1, Dnmt3a, and Dnmt3b mRNA in group B were the highest, and those in group C were the lowest. The expression levels of tumor suppressor genes p16 and RASSF1A mRNA in group B were the lowest, and the highest were in group C. Blocking ERK pathway could inhibit Dnmts expressions with increasing expression levels of tumor suppressor genes p16 and RASSF1A. The same conclusions were obtained through a study on the protein expression levels of Dnmt1, Dnmt3a, Dnmt3b, p16, and RASSF1A mRNA through Western blot. The Dnmts expression levels of tumor cells were significantly higher than those of normal cells and related to high methylation status of promoters in tumor suppressor genes. Through the detection of Dnmts mRNA levels in renal cell carcinoma through semi-quantitative PCR, Dnmt1 and Dnmt3b mRNA expression levels were found to significantly increase but eventually significantly descended and presented high methylation status through the detection of relevant tumor suppressor genes. In addition, Dnmt1 mRNA expression level had a correlation with prognosis. p16 is a commonly seen tumor suppressor gene, and studies on its methylation status have been carried out in a large quantity. The studies found that p16 was silenced due to the methylation of its CpG islands and the coding function of protein level declined or closed that led to tumor genesis. As a tumor suppressor gene, RASSF1A participates in ERK signal pathway, which results in the transduction pathway of growth effect signals. The following effects were observed. Tumor cell proliferation was inhibited. The apoptosis and senescence of tumor cells were facilitated by the inhibition of the activation of Ras kinases. Thus, the functional inactivation of RASSF1A results in failure of inhibiting effect on tumor cells. Researchers have detected the expression levels of p16 and RASSF1A mRNA in non-small cell lung carcinoma and found that their expression levels are distinctly down-regulated [6,7]. Therefore, blocking ERK signal pathway, down-regulating Dnmts and its protein expression levels, and up-regulating p16 and Ras mRNA and its protein expression levels can exert an inhibitory effect on the proliferation and growth of tumor cells.
Through the detection of the methylation status of the promoter regions of p16 and RASSF1A, the methylation status of tumor suppressor genes p16 and Ras in group B was found to be distinct. P16 in group C presented unmethylated status, and RASSF1A presented partial methylation status and partial demethylation status. This test indicated that inhibiting ERK pathway could facilitate the methylation of tumor suppressor genes p16 and RASSF1A and activating ERK pathway could inhibit their methylation. Therefore, possible regulatory mechanism of ERK signal transduction pathway for DNA methylation level was Ras activated. Ras/ERK in tumor cells mediated the methylation of tumor suppressor genes CpG islands by up-regulating Dnmts gene expression. By contrast, if ERK signal transduction pathway activated by Ras was blocked, then Dnmts expression levels would be downregulated to block the mediating effect of this signal transduction pathway on methylation. Due to the methylation of the promoter regions in multiple tumors, p16 genes were silenced with inactivated expression. The study on RASSF1A also indicated that its main inactivation mechanism was the methylation of CpG locus in the promoter region and taken as an alternative tumor suppressor gene for lung carcinoma [8,9]. Tumor cells with significantly increasing protein expression levels of Dnmts show significant increase in methylation degrees in the promoter regions of tumor suppressor genes p16 and RASSF1A. In addition, blocking the ERK signal transduction pathway can inhibit DNA methylation effect and activate tumor suppressor genes p16 and RASSF1A to inhibit cancer cell proliferation [10].
Conclusion
Results of our study on the regulatory mechanism of ERK signal pathway for DNA methylation of lung carcinoma cells showed that blocking ERK signal pathway leads to the inhibition of cell proliferation rate and DNA methylation level. It also inhibits proliferation and growth of cancer cells by down-regulating Dnmts mRNA and their protein expression levels and by up-regulating tumor suppressor genes p16 and RASSF1A mRNA and their protein expression levels.
Acknowledgements
This work was supported by grants from the Key Technologies Research and Development Program of Guangxi (No. 14124004-1-4).
References
- Wang H, Zhang Y, Qiao M. Mechanisms of extracellular signal-regulated kinase/cAMP response element-binding protein/brain-derived neurotrophic factor signal transduction pathway in depressive disorder. Neural Regen Res 2013; 8: 843-852.
- Yang A, Peng H, Huang Z, Wu H, Wu L. Regulation of osteosarcoma proliferation and apoptosis by miR-489 through targeting SOX4 gene. Biomed Res India 2017; 28: 4282-4286.
- Dine J, Ducourneau VRR, Fenelon VS, Fossat P, Amadio A, Eder M, Israel JM, Oliet SHR, Voisin DL. Extracellular signal-regulated kinase phosphorylation in forebrain neurones contributes to osmoregulatory mechanisms. J Physiol 2014; 592: 1637-1654.
- Cai DT, Zhang YB, Chen K, Xiong QX, Luo WJ, Gao ZG. Synthesis and Evaluation of a Novel Heterocyclic Compound against Pediatric Hepatoblastoma Cells. Lat Am J Pharm 2017; 36: 2022-2027.
- Richardson ET, Shukla S, Sweet DR, Wearsch PA, Tsichlis PN, Boom WH, Harding CV. Toll-like receptor 2-dependent extracellular signal-regulated kinase signaling in mycobacterium tuberculosis-infected macrophages drives anti-inflammatory responses and inhibits Th1 polarization of responding T cells. Infect Immun 2015; 83: 2242-2254.
- Zhao S, Teng F, Li P, Gao F, Wu Q. The anti-cancer activity of astragaloside on human breast cancer cells in vitro. Lat Am J Pharm 2017; 36: 2037-2041.
- Wang Y, Han R, Zuo Z. Dexmedetomidine post-treatment induces neuroprotection via activation of extracellular signal-regulated kinase in rats with subarachnoid haemorrhage. Br J Anaesth 2016; 116: 384-392.
- Yang Y, Zhou JY, Zhao LJ, Gao BR, Wan XP, Wang JL. Dual-specificity phosphatase 1 deficiency induces endometrioid adenocarcinoma progression via activation of mitogen-activated protein kinase/extracellular signal-regulated kinase pathway. China Med J (Engl) 2016; 129: 1154-1160.
- Sun J, Nan G. The extracellular signal-regulated kinase 1/2 pathway in neurological diseases: A potential therapeutic target (Review). Int J Mol Med 2017; 39: 1338-1346.
- Liu MK, Brownsey RW, Reiner NE. Gamma interferon induces rapid and coordinate activation of mitogen-activated protein kinase (extracellular signal-regulated kinase) and calcium-independent protein kinase C in human monocytes. Infect Immun 1994; 62: 2722-2731.