ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2009) Volume 20, Issue 2
Prevalence of Mec A Gene positive coagulase negative Staphylococci in NICU of a tertiary care hospital
1P.G. Department of Microbiology, Chhatrapati Shahuji Maharaj Medical University, Lucknow, India
2Department of Pediatrics, Chhatrapati Shahuji Maharaj Medical University, Lucknow, India
3Department of Physiology,, Chhatrapati Shahuji Maharaj Medical University, Lucknow, India
4Department of Biochemistry, Chhatrapati Shahuji Maharaj Medical University, Lucknow, India
- *Corresponding Author:
- R.K. Singh
Department of Biochemistry
Chhatrapati Shahuji Maharaj
Medical University (Formerly King George’s Medical University) Lucknow, India.
Accepted February 03 2009
For last 20 years Coagulase negative staphylococci (CoNS) have been identified as major causes of late-onset neonatal bacteraemia in new born intensive care units. In our study an attempt was made to evaluate prevalence of oxacillin resistance CoNS in NICU. The study group comprised of 1087 cases admitted in the NICU of the Gandhi Memorial and associated hospitals, Lucknow during the period of September 2005 to December 2007. Cases included in study were neonates (of age less than or equal to 28 days) suspected to be suffering from the septicemia on the clinical grounds. Samples taken were blood, pus, nasal aspirate, urine and catheter tip. Biotyping was done for all the CoNS isolates. In biotyping S. epidermidis (40%) was the most common species followed by S. saprophyticus (20%) and S. haemolyticus (12%). 54% CoNS were found to be methicillin resistance by DD(oxacillin) and 52% isolates were positive for mecA gene. CoNS causing neonatal septicemia are resistant to the routine antibiotics used to treat newborn infants and vancomycin is often required for adequate therapy.
Keywords
Coagulase negative staphylococci (CoNS), neonatal bacteraemia, oxacillin resistance, biotyping, septicemia
Introduction
For last 20 years Coagulase negative staphylococci (CoNS) have been identified as major causes of late-onset neonatal bacteraemia in new born intensive care units [1,2]. Babies with CoNS infection are smaller and more premature than controls but even after controlling for prematurity, they stay longer in neonatal intensive care units than non-infected babies and at considerable cost CoNS infections are associated with a significant mortality and morbidity [3,4] including an increase in the duration of hospital stay and health care costs in adults [5] and VLBW infants [6]. CoNS were considered non pathogenic until their implication as a nosocomial agents and emergence of multiresistance [7]. Healthcare associated infections caused by these microorganisms are responsible for high mortality rates worldwide. The emergence of CoNS population with heterogenous resistance to oxacillin causes a great difficulty to detect them in clinical routine laboratories [8]. In Brazil oxacillin resistance may be present in over 80% of isolates in some health institutions [9]. On this account vancomycin has been widely used and it is major cause for the emergence of glycopeptide resistant isolates [10,11]. Our aim of the study was to show the prevalence of methicillin resistant CoNS in NICU of our teaching hospital.
Material and Methods
Study plan
The study was carried out in the P.G. Department of Microbiology, Chhatrapati Shahuji Maharaj Medical University, Lucknow, India. Neonates with signs and symptoms of septicemia attending the Department of Paediatrics, Gandhi Memorial and Associated Hospitals from Sept., 2006 to Dec. 2007 were included in this study.
All the samples were subjected to Grams staining. For identification of CoNS was done by catalase test, coagulase test, novobiocin sensitivity, O/F test, acetoin production, phosphatase test, sugar fermentation test, ornithine decarboxylase test and urease test. For positive samples of CoNS antimicrobial susceptibility test was done by Kirby-Bauer disk diffusion method.
Antimicrobial susceptibility test
This test was performed on Mueller – Hinton agar with the following antibiotic discs (Hi Media Laboratories) using the Kirby- Bauer disc diffusion method (DD): penicillin (10 U), cefotaxime (30 μg), vancomycin (30 μg), teicoplanin (30 μg), C/S (75/30 μg), Ceftriaxone(30 μg), gentamycin (10 μg), amikacin (30 μg) and ciprofloxacin (5 μg). An oxacillin disc (1 μg) was used to detect methicillin resistance. Sensitivity was read after incubation for 24 h at 35° C. Isolates were regarded as sensitive or resistant according to CLSI criteria (CLSI 2005) [12].
DNA amplification
A 310 bp fragment of mec A was amplified with primers
M1= TGGCTATCGTGTCACAATCG
M2= CTGGAACTTGTTGAGCAGAG[13]
250μM of each deoxynucleotide tri phosphate, 20 pM of each primer, 1.1U DNA Tth Plus DNA polymerase and 10μL of DNA, made up to volume of 50μL in Milli Q water and overlaid with 40 μL of paraffin oil. Amplification was performed in perkin Emler DNA Tharmal Cycler 480. The initial denaturation step at 92°C for 3 minutes, was followed by 30 cycles consisting of denaturation at 92°C for 1 minute, then a final extension step at 72°C for 3 minute. Mec A positive isolates generated at 310 bp fragment, which was detected by agrose gel electrophoresis of the amplified product.
Observations and Results
The study group comprised of 1087 cases admitted in the NICU of the Gandhi Memorial and associated hospitals, Lucknow during the period of September 2005 to December 2007. Cases included in study were neonates (of age less than or equal to 28 days) suspected to be suffering from the septicemia on the clinical grounds. All the cases were subjected to blood culture examination. Isolates were identified and tested for antimicrobial susceptibility. Samples collected were blood, pus, nasal aspirate, urine and catheter tip.
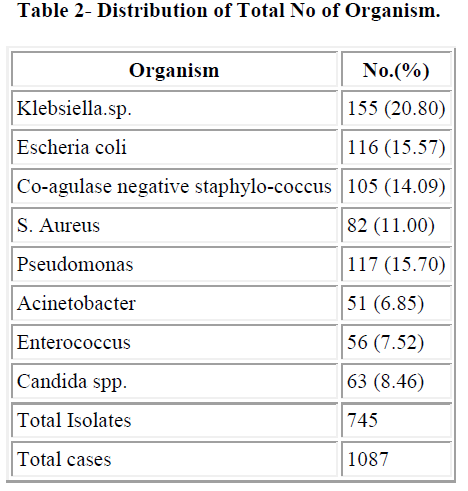
The maximum numbers of samples were blood samples in all three years followed by catheter tip. We got maximum number of blood samples in year 2006 (70.07%) followed by 2007 (69.45%). Distribution of total isolates are illustrated in table 2. In this study Co-agulase negative staphylococcus (14.09%) and S. aureus (11%) were most common gram positive isolates and Klebsiella.spp (20.8%), E.coli (15.57%) and Pseudomonas spp (15.70%) were most common gram negative organism.
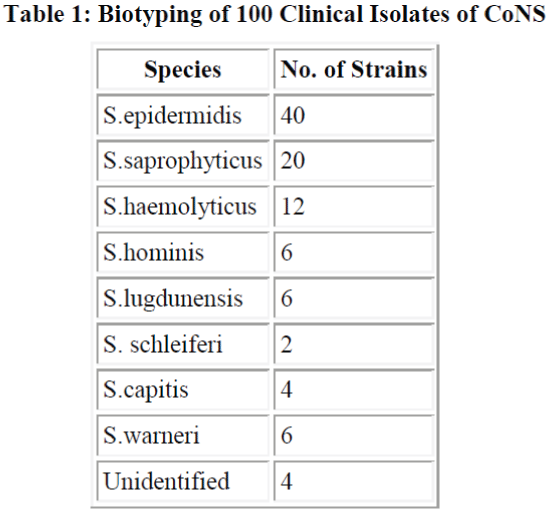
Total 100 samples of CoNS were biotyped S. epidermidis (40%) was the most common species followed by S. saprophyticus (20%) and S. haemolyticus (12%) (Table 1). Sixty out of 100 strains of CoNS (60%) were isolated from blood samples, 24 from pus samples, 10 from urinary catheter tip and 6 from the urine samples. All 100 strains were negative for blood clumping factor and tube coagulase negative. Among CoNS resistance was seen maximum with penicillin (80%). Resistance was minimum with amikacin which was 21.6%. We found 54% of strains were resistant to oxacillin and compared oxacillin resistant and oxacillin sensitive CoNS strains for other antibiotics.
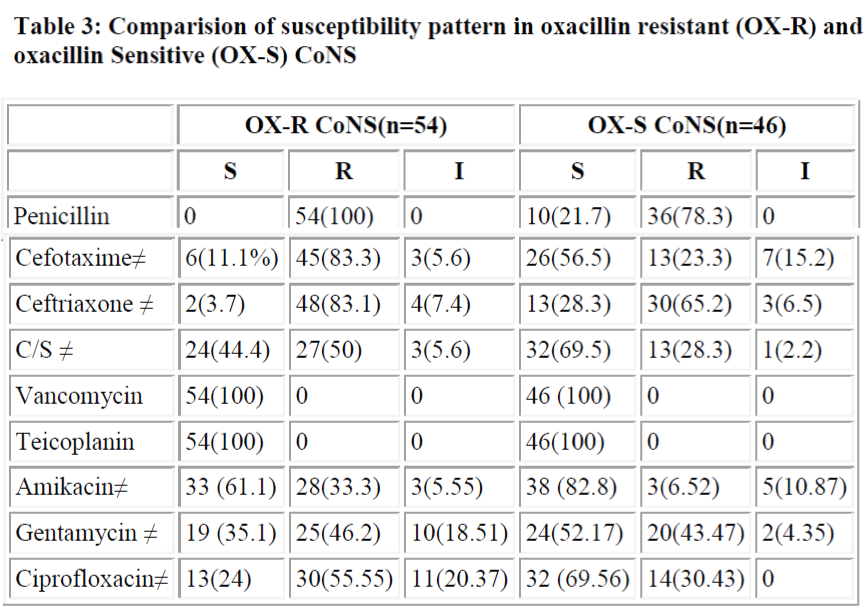
All oxacillin resistant or sensitive strains were sensitive to vancomycin and teicoplanin, but resistance to other drugs in oxacillin resistance stains were more common. Resistance to all antibiotics is significantly more in oxacillin resistant group except for gentamycin (P>0.05) (table 3). Oxacillin disk diffusion and PCR screened 54% and 52% mec A positive CoNS.
Discussion
Prevalence of CoNS septicemia has increased in the past decades. Similarly, rate of multiple drug resistant CoNS (MRCoNS) has also increased, and administration of first dose of antibiotic, which is started without waiting for culture report, is directed towards Gram negative organism. In present study we found that Candida spp. accounted for 8.46% of septicemia. In a previous study at our place Roy et al. [14] have reported 6.8% cases of candidal septicemia. In a study by Rani et al. [15], revealed 34.7% of candidal septicaemia. Usha Arora and Pushpa Devi [16] have reported 4.51% Candida spp. from the cases of septicemia. In our study Klebsiella spp. (20.30%) was predominant followed by E. coli (15.57%), Pseudomonas spp. (15.70%), CoNS (14.09%) and S. aureus (11%). Shahsanam Gheibi [17] found 33% Pseu- domonas spp., 22% Klebsiella spp., 5% Enterobacter and 5% E. coli and CoNS was responsible for 27% of Noso- comial infection. Nalini et al. [18] found S. aureus (35.3%) as a predominant organism followed by Klebsiella spp. (18.37%). Prevalence of CoNS septicaemia has 14.09% which was slightly lower than previous study done by Roy et al. (2002) [14]. In India and abroad different authors have reported different incidence of rates of CoNS septicemia ranged from 2.3% to 55%. Mahapatra et al. (2002) [19], Sharma et al. (2002) [20], Baungart et al. Leon et al.[22] have reported 2.3%, 2.87%, 12% and 11% incidence of CoNS septicemia respectively. Similarly, Chaturvedi, Agarwal and Narang [23], Berger et al. [24] and Mokoulu AO, Jiya N and Adesiyun OO [25] isolated 24%, 27% and 24.6% CoNS respectively in their study, which is comparable to present study, while Sanghvi and Tudehope [26], Hansey OJ [27] have reported higher incidence rate, 38.8% 49% and 55% respectively Among the Gram negative group, Enterobacteriacae family predominated, Klebsiella spp. (20.80%) was most common. Other authors also reported Klebsiella spp. as most common gram negative organism (Nalini et al., 2004; Rani et al., 2004) [18,15] while many other have reported Klebsiella as most common cause of neonatal septicemia including the previous study done at our place by Roy et al. [14]. They have reported 24.6% cases of neonatal septicemia caused by Klebsiella spp. National Neonatal Perinatal Data base (2002-03) have also reported an incidence of 32.5% of Klebsiella septicemia. Sharma et al. [20] have reported even higher rate, 61.4% whereas Shahsanam et al. reported 22% of Klebsiella septicemia. Mahapatra et al.[19]
found that Enterobacter (39.5%) was most common organism causing neonatal septicemia. They also reported 4.6% incidence of citrobacter septicemia. E. coli was isolated less frequently (3.7%) in present study compared to the previous study (14%). We biotyped 100 strains of CoNS irrespective of their significance or insignificance and found that most common species was S. epidermidis (40%) followed by S. saprophyticus (20%) and S. haemolyticus (12%). Other species identified were S. homini’s (6%), S. lugdunensis (6%), S. schleiferi (2%), S. capitis (4%) and S. warneri (6%). Raimundo et al. [28] isolated 55 CoNS from the blood of neonates in a NICU. Most common species were S. epidermidis, S. haemolyticus and S. warnei. In other study Goyal et al.[29] observed S. epidermidis, S. haemolyticus and S. saprophyticus and S. horminis upto the species level which is almost similar to our study.
A study by Fluit et al. [30] showed that 100% of MRSA and MRCoNS were penicillin resistant while in methicillin sensitive strains the figures were comparatively lower (84.9% in MSSA and 72.8% in MS CoNS). In our study also 100% of MRCoNS were penicillin resistant, while only 78.3% of MSCoNS showed resistance to penicillin. More than 80% were resistant to third generation cephalosporins. Co-existing resistance to different antibiotics (except gentamycin) is significantly higher in MRCoNS in comparison to MSCONS (p<0.05) (Table 4). None of the strains in our study showed resistance to Vancomycin and teicoplanin in vitro and these drugs therefore can be effectively used if methicillin resistance suspected drug treatment.
As the majority of methicillin resistant Staphylococci harbour the mec A gene, test based on amplification such as PCR have identified even the most heterogenous strains correctly [31]. Although PCR is considered the best method for detection of oxacillin resistant Staphylococci.
References
- Report of the National Neonatal Perinatal Database (National Neonatology Forum) 2002-2003.
- Gaynes RP, Edwards JR, Jarvis WR, et. al. Nosocomial infections among neonates in high-risk nurseries in the United States. Pediatrics.1996; 98:357-361.
- Stoll BJ, Hansen N, Fanaroff AA, et. al. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics., 2002; 110: 285-291.
- Karlowicz MG, Buescher ES, Surka AE. Fulminant late onset sepsis in a neonatal intensive care unit, 1988-1997, and the impact of avoiding empiric vancomycin therapy. Pediatrics. 2000; 106:1387-1390.
- Pittet D, Tarara D, Wenzel RP. Nosocomial bloodstream infection in critically ill patients. Excess length of stay, extra costs, and attributable mortality. JAMA 1994; 271: 1598-1601.
- Gray JE, Richardson DK, McCormick MC, et. al. Coagulase-negative staphylococcal bacteremia among very low birth weight infants: relation to admission illness severity, resource use, and outcome. Pediatrics 1995; 95: 225- 230.
- Beekmann SE, Diekema DJ, Chapin KC. Effects of rapid detection of bloodstream infections on length of hospitaliza- tion and hospital charges. J Clin Microbiol 2003; 41: 3119-3125.
- Cauwelier B, Gordts B, Descheemaecker, Van Landuyt H. Evaluation of a disk diffusion method with cefoxitin (30 µg) for detection of methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol 2004; 6: 389-392.
- Sader HS, Gales AC, Pfaller MA, Mendes RE, Zocolli C, Barth AL, Jones RN. Pathogen frequency and resistance patterns in Brazilian hospitals: summary of results from three years of the SENTRY Antimicrobial Surveillance Program. Braz J Infect Dis 2001, 5: 200-214.
- Oliveira, GA, Faria, JB, Levy, CE, and Mamizuka, EM. Characterization of the Brazilian endemic clone of methicillin-resistant Staphylococcus aureus (MRSA) from hospitals throughout Brazil. Braz. J. Infect. Dis. 2001; 5:163–170.
- Nunes APF, Teixeira LM, Iorio NLP, Bastos CCR, Fonseca LS, Souto-Padrón T, Santos KRN. Heterogeneous resistance to vancomycin in Staphyloccus epidermidis, Staphylococcus haemolyticus and Staphylococcus warneri clinical strains: characterization of glycopeptide susceptibility profiles and cell wall thickening. Int J Antimicrob Agents. 2006; 27: 307-315.
- Clinical and Laboratory Standard Institute. Performance standards for antimicrobial disk susceptibility tests, 15th ed. Vol. 25, no. 1. Approved standard N2- A8. Clinical and Laboratory Standard Institute 2005; Wayne, Pa.
- Vannuffel, P,Gigi J, Ezzedine H, Vandercam B, Delmee M, Wanters G and Gala JL. Specific detection of methicillin resistant Staphylococcus species by multiplex PCR. J. Clin Microbiol 1995; 33:2864-2867.
- Roy I, Jain A, Kumar M, agarwal SK. Bacteriology of neonatal septicemia in a tertiary North Indian Hospital. Indian J of Medical Microbiol 2002; 20(3):156- 159.
- Rani R, Mahapatra NP, Mehta G, Randhawa VS. Changing trends of candida species in neonatal septicaemia in a tertiary North Indian Hospital. Indian J of Medical Microbiol 2002; 20(1):42-44.
- Usha A, Pushpa D. Bacterial profile of blood stream infections and antibiotic resistance pattern of isolates. JK Science 2007; 9 (4): 186-190.
- Shahsanam G, Zahra F, Karamyyar M, Khahabi J. Coagulase negative staphylococcus, the most common cause of neonatal septicemia in Urmia, Iran. Iran J Pediatr 2008; 18 (3); 273-243.
- Nalini et al. Antimicrobial susceptibility of isolates from neonatal septicemia; Jpn J Infect Dis 2004; 57, 273-275.
- Mahapatra A, Gosh SK, Mishra S, Pattnaik D, Pattnaik K, Mohanty SK. Enterobacter cloacal: a predominant pathogen in neonatal septecimia. Ind J of Med Microbiol 2002; 20 (2): 110-112.
- Sharma M, Goel N, Chaudhary U, Agarwal R, Arora DR. Bacteremia in children. Indian J. Pediat- rics.Dec.2002; 69: 1029- 032.
- Baumgarth S, Susan EH, Joseph MC, Richard AP. Sepsis with coagulase negative staphylococci in critically ill newborns. Am J Dis Child 1983; 137: 461-463.
- Leon SP De, Wenzel RP. Hospital aquired blood stream infections with Staphylococcus epidermidis. American J of Medicine 1984; 77: 639-644.
- Chaturvedi P, Agarwal M, Narang P.Analysis of blood -culture isolates from neonates of a rural hospital. Indian Pediatr1989; 26 (5): 460-465.
- Berger A, Salzer HR, Weninger M, Segeder B, Aspock C. Septicemia in Austrian intensive care unit: a 7 year analysis. Acta Paediat 1998; 87(10):1066-9.
- Mokuolu AO, Jiya N, Adesiyum OO. Neonatal septicemia in Ilorin: bacterial pathogens and antibiotic sensitivity pattern. Afr J Med Sci 2002 June; 31(2):127- 130.
- Sanghvi KP, Tudehope DI. Neonatal bacterial sepsis in neonatal intensive care unit: a 5 years analysis. J Pediatr Child Health 1996; 32 (4): 333-338.
- Hensey OJ, Hart CA, Cook RW. Serious infection in neonatal intensive care unit: A two years survey. J. Hyg (lond) 1985; 95(2): 289-97.
- Raimundo O, Heussler H, Bruhn JB, Suntrarachum S, Kelly N, Deighton MA, Garland SM. Molecular epidemiology of coagulase-negative staphylococcal bacteraemia in new born intensive care unit. J.Hospital Infect 2002; 51 (1): 33-42.
- Goyal R, Singh NP, Kumar A, Kaur I, Singh M, Sunita N, Mathur M. Simple and economical method for speciation and resistotyping of clinically significant coagulase negative staphylococci, Indian J. Med. Microbiol 2006; 24: 201-204.
- Fluit AC, Jones ME, Schmitz, FJ, Acar J, Gupta R, Verkoef J. Antimicrobial susceptibility and frequency of occurrence of clinical blood isolates in Europe from the SENTRY Antimicrobial Surveillance Programme, 1997 and 1998. Cl in Infect Dis 2000; 30, 454 – -4 6 0.
- Jaffe RI, Lane JD, Albury SV and Niemeyer DM. Rapid extraction from a direct identification in clinical samples of methicillin resistant staphylococci using PCR. J Clin Microbiol.2000; 38; 3407-3412.