ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 8
Phyllanthus emblica L. extract activates Nrf2 signalling pathway in HepG2 cells
1College of Pharmacy, Liaoning University of Traditional Chinese Medicine, Dalian, PR China
2Liaoning Grand Nuokang Biopharmaceutical Co. Ltd, Shenyang of Liaoning Province, PR China
- *Corresponding Author:
- Hongying Wang
Liaoning Grand Nuokang Biopharmaceutical Co. Ltd
Shenyang of Liaoning province, PR China
Accepted on October 31, 2016
To investigate the effects of Extracellular signal-Regulated Kinase (ERK), p38 mitogen-Activated Protein Kinase (p38MAPK) and other kinase pathways on the activation of Nuclear related factor 2 (Nrf2) signalling pathway by Phyllanthus emblica L. extract (PET). After pre-treatment with different protease inhibitors for 2 h, cells were interfered with Phyllanthus emblica L. extract for 24 h. mRNA expressions of Nrf2 and downstream target genes HO-1, NQO1, P-gp, MRP2 were detected by RT-PCR. Protein level and nuclear translocation of Nrf2 were detected by Western blotting. RT-PCR results showed that treatment with PD98059, SB203580 and rottlerin suppressed the HO-1, NQO1, P-gp and MRP2 mRNA induction by PET, but only treatment with SB203580 and rottlerin could inhibit the Nrf2 mRNA expression by the extract. Western blotting showed that treatment with PD98059, SB203580 and rottlerin suppressed the HO-1 and P-gp protein inducing effect of PET. Treatment with various inhibitors could all induce the cytoplasmic expression of Nrf2 protein, but treatment with PD98059 and SB203580 could inhibit nuclear translocation of Nrf2. Nrf2 pathway activating mechanisms of PET may be associated with the roles of ERK and p38MAPK in directly phosphorylating Nrf2 and promoting Nrf2 nuclear import to increase its nuclear accumulation.
Keywords
Phyllanthus emblica L. extract, Nuclear related factor 2 (Nrf2), Extracellular signal-regulated kinase (ERK), p38 mitogen-activated protein kinase (p38MAPK), Signalling pathway, Liver hepatocellular cells (HepG2).
Introduction
Nuclear related factor 2 (Nrf2) is a novel protein found by Moi et al. during cloning experiment in 1994. Due to a high degree of homology with nuclear factor E2, the name was given as Nrf2. Nrf2 is the most dynamic transcription factor among basic leucine zipper Cap'n'Collar family members, which is a key factor regulating the bodies or cells to antagonize extraneous matters and oxidative damage [1]. Studies have shown that Nrf2 plays an important role in numerous pathophysiological processes, which is a vital regulatory factor resisting oxidative stress discovered recently. Nrf2 regulates the transcriptional activity of downstream target genes such as ARE-dependent phase II detoxification enzymes by acting with Antioxidant Response Element (ARE) as a transcription factor [2]. When organisms or cells are stimulated by reactive oxygen, electrophilic substances or upstream signalling pathways (kinase pathway, etc.), Nrf2 is activated and dissociated from cytoplasmic adapter protein (Keap1) into nuclei, thereby binding with ARE to initiate the transcription of downstream target genes [3].
The present study revealed that Protein Kinase C (PKC), Mitogen-Activated Protein Kinase (MAPK) and other protein kinases can directly phosphorylate Nrf2 and dissociate it from Keap1 to activate Nrf2 and induce transcription of downstream target genes. Studies reported that PKC and extracellular signal-regulated kinase (ERK) may up-regulate Nrf2 by increasing the transcriptional activity of Nrf2, thereby up-regulating the expression of phase II enzyme glutathione synthetase (γ-GCS) [4]. Wu et al. found that after giving p38 mitogen-activated protein kinase (p38MAPK) and ERK pathway inhibitor, respectively, expression of Heme Oxygenase-1 (HO-1) in lungs of endotoxin shock rats was down-regulated [5,6]. Zixiang et al. found that ERK signalling pathway inhibitor PD98059 could inhibit the nuclear translocation of Nrf2 and significantly reduce the intra-nuclear protein level of Nrf2 [7].
PET is the ethanol extract of Phyllanthus emblica L., whose primary active constituents are polyhydroxyphenols [8]. Our preliminary study showed that PET significantly induced the nuclear translocation of Nrf2 and the transcription of downstream target genes HO-1, P-glycoprotein (P-gp) and other phase II detoxifying enzymes and transporters through Nrf2 signalling pathway.
In this study, HepG2 cells were pre-treated with different protease inhibitors and then interfered with PET. Intracellular expressions of HO-1, P-gp and other phase II detoxifying enzymes and transporters were observed. Meanwhile, Nrf2 expression and changes in its nuclear translocation were detected, and the effects of different kinase pathways on PET activation of Nrf2 signalling pathway were investigated, which were reported below.
Materials and Methods
Cell lines: HepG2 cell lines were purchased from the Cell Resource Center of the Shanghai Institutes for Biological Sciences, CAS.
Reagents and instruments: DMEM high glucose medium (purchased from GIBCOBRL); penicillin+streptomycin (purchased from Sigma Chemical Co.); 2.5 g/L trypsin (purchased from GIBCOBRL); FBS (purchased from Invitrogen Corporation); MTT (purchased from Roche); dimethyl sulfoxide (purchased from Sigma Chemical Co.); Trizol kit, RT-PCR kit and PCR amplification kit were purchased from TaKaRa, Japan. BDS200 inverted microscope (Nikon, Japan); incubator (Thermo, USA); nucleic acid protein detector (Thermo, USA); PCR instrument (ABI, USA).
Drugs and preparation
LY294002 (MedChem Express, USA); PD98059 (MedChem Express, USA); SB203580 (MedChem Express, USA); Rottlerin (Sigma, USA); crude Phyllanthus emblica L. (Dalian Baidu Medicine Co., Ltd.; batch No.: YPA2L0001; identified by Professor Zhang of the Medicinal Chemistry Department, School of Pharmacy, China Medical University as the dried ripe fruit of Phyllanthus emblica L. in the genus Phyllanthus of the family Euphorbiaceae. PET was reflux extracted with ethanol using crude Phyllanthus emblica L. and purified through D101 macroporous resin, where the content of polyhydroxyphenol compounds was 39.77 mg/g.
Cell culturing
HepG2 cells were cultured in DMEM high glucose medium containing 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin, cultured routinely at 37°C, 5% CO2 with saturated humidity and digested and passaged with 2.5 g/L trypsin (containing 2 g/L EDTA).
Drug treatment and grouping
Experiment was divided into blank control group, PET treated group, PET+PD98059 (ERK specific antagonist) treated group, PET+LY294002 (PI3K/Akt specific antagonist) treated group, PET+SB203580 (p38MAPKs specific antagonist) treated group and PET+Rottlerin (PKC specific antagonist) treated group. Blank control group was inoculated only with cells and added with an isovolumetric incomplete DMEM high glucose medium. After pre-treating with protein kinase inhibitor for 2 h, HepG2 cells were treated with PET for 24 h then collected.
QRT-PCR detection of HO-1, NQO1, P-gp, MRP2 and Nrf2 mRNA expressions
Cells were collected, total cellular RNA was extracted according to kit instructions, and 1 μg of total RNA was reverse transcription reacted using RT-PCR kit. According to RT-PCR kit instructions, semi-quantitative analysis was performed on cellular Nrf2, HO-1, NQO1, P-gp and MRP2 mRNA expressions. RT-PCR cyclic conditions were: pre-denaturation at 95°C for 30 s; denaturation at 95°C for 15 s; annealing at 59°C for 30 s; extension at 72°C for 31 s; a repeat of 40 cycles; and a final extension at 72°Cfor 5 min. Results were processed by 2-Ct method, where target gene Ct=target gene Ct-reference gene Ct; target gene Ct=target gene Ct-reference gene Ct.
Western blotting assay of HO-1, P-gp and Nrf2 protein levels
Cells were collected, and total protein and nucleoprotein were extracted. After centrifugation at 4°C, 12000 g for 5 min, supernatant was collected, and total cytoplasmic protein was determined by BCA method [9]. Equal amounts of protein were collected from each group for 120 g/L SDS-PAGE gel electrophoresis, transferred to membranes and closed for 2 h. Membranes were soaked in diluents containing primary antibodies (HO-1, P-gp and Nrf2 antibody concentrations of 1:2,000), incubated overnight at 4°C, washed sufficiently with Tween-20 Phosphate Buffer Solution (PBST solution) for 5 min five times. Membranes were soaked in diluents containing secondary antibody (antibody concentration of 1:2,000), incubated at room temperature for 2 h, then washed and developed with chromogenic agent. The experiment was repeated three times. Protein level percentage of each sample phase relative to the control group was calculated with blank control group as 100%, and statistical analysis was performed. Target protein expression=target protein value/internal reference Glyceraldehyde Phosphate Dehydrogenase (GAPDH) value.
Statistical methods
Experiments were repeated three times. Data were processed using SPSS 16.0 software, and results were expressed as x ± s. Inter-group comparisons were performed by one-way ANOVA, and significance level was set at α=0.05.
Results
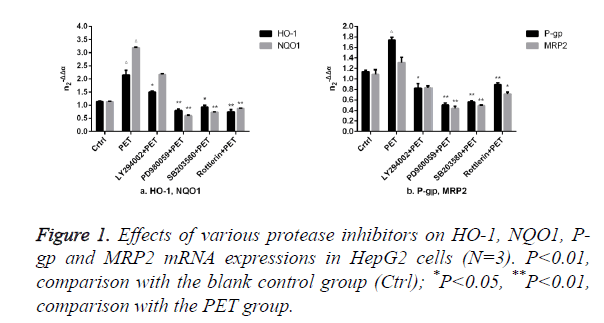
Effects of protease inhibitors on mRNA expressions of HO-1 and other Nrf2 downstream target genes in HepG2 cells
The results of Figure 1 showed that compared to the blank control group (ctrl), PET group could significantly induce the mRNA expression. After pre-treatment with specific protease inhibitor, the induction effect weakened significantly. Compared to PET group, treatment with PD98059, SB203580 and Rottlerin could suppress the HO-1, NQO1, P-gp and MRP2 mRNA inducing effect of PET, with PD98059 and SB203580 exhibiting the most prominent effects.
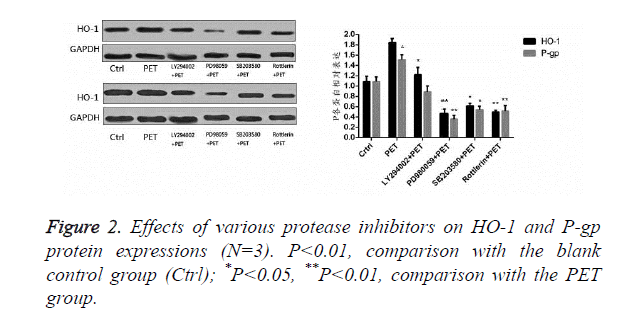
Effects of protease inhibitors on HO-1 and P-gp proteins in HepG2 cells
According to the results of Figure 2, after pre-treatment with different protease inhibitors and subsequent intervention with PET, detection of HO-1 and P-gp protein expression levels in HepG2 cells found that PET could significantly induce HO-1 and P-gp protein expressions compared to the blank control group. After treating with kinase inhibitor, the induction effect disappeared and was inhibited to varying degrees, with PD98059 exhibiting the most prominent inhibitory effect on HO-1 and P-gp proteins. Compared to the PET group, various inhibitor treated groups could all differentially inhibit the HO-1 and P-gp protein expressions.
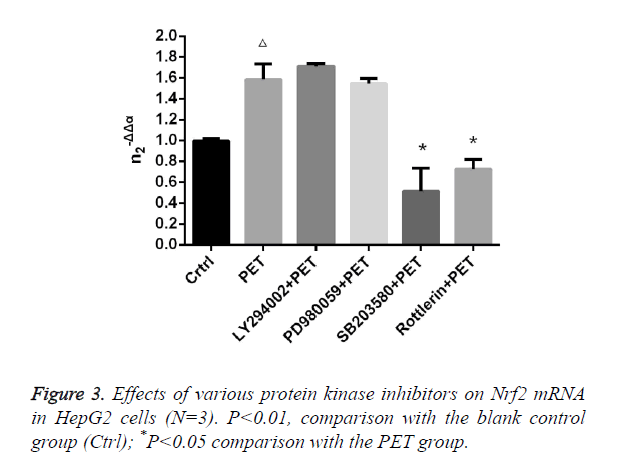
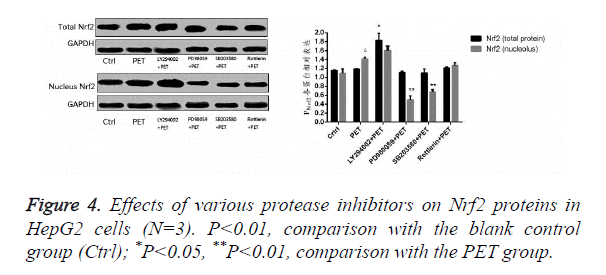
Effects of various protease inhibitors on Nrf2 expression level and nuclear translocation (Figures 3 and 4)
According to the results of Figure 3, after pre-treatment with different protease inhibitors and subsequent intervention with PET, detection of Nrf2 mRNA expression in HepG2 cells found that both LY294002 and PD98059 could induce Nrf2 mRNA expression to varying degrees compared to the blank control group. SB203580 and Rottlerin, on the other hand, exhibited inhibitory effect, with an up to 50% inhibition rate for SB203580. Compared to the PET group, only SB203580 and Rottlerin could inhibit the Nrf2 mRNA inducing effect of PET. According to the results of Figure 4, after pre-treatment with different protease inhibitors and subsequent intervention with PET, detection of total cytoplasmic protein in HepG2 cells found that all groups could induce intracytoplasmic Nrf2 protein expression to varying degrees compared to the blank control group, with LY294002 exhibiting the strongest effect. Detection of nucleoprotein found that only PD98059 and SB203580 had Nrf2 inhibiting effect. Compared to the PET group, LY294002 could induce cytoplasmic Nrf2 protein expression, while PD98059 and SB203580 could suppress nuclear import of Nrf2.
Discussion
Phyllanthus emblica L. is a plant in the genus Phyllanthus of the family Euphorbiaceae [10], which is widely distributed in tropical and subtropical regions. Domestically, Phyllanthus emblica L. is distributed mainly in Yunnan, Guangxi, Guangdong, Fujian, Hainan, Taiwan, Sichuan and Guizhou, of which Fujian and Yunnan are the largest producers [11]. Sour and bitter in taste with sweet aftertaste, Phyllanthus emblica L. is a wild plant resource of high edible and medicinal values. It has unique flavour, rich nutrition and strong healthcare function and contains a variety of beneficial active substances, which has tonic, anti-tumor, anti-aging, anti-inflammatory, anti-bacterial, anti-viral, anti-mutagenic, anti-hypertension and lipid regulating effects [12]. Our previous study found that PET could significantly induce Nrf2 nuclear translocation, negatively regulate Keap1 protein expression while inducing transcription of HO-1, P-gp and other phase II detoxifying enzymes and transporters through Nrf2 signalling pathway. Existing research suggests that many kinase pathways, such as Protein Kinase C (PKC), Mitogen-Activated Protein Kinase (MAPK) and Phosphatidylinositol 3-Kinase (PI3K), are involved in the activation of Nrf2 [13]. In this study, the role of kinase pathways in PET activation of Nrf2 pathway was investigated by administering SB203580, PD98059 and other protease inhibitors. The results found that after pre-treatment with protease inhibitors, FSE's ability to induce downstream target genes HO-1 and P-gp reduced markedly, with inhibitors PD98059 and SB203580 exhibiting the most obvious effects. In addition, as PD98059 and SB203580 were specific inhibitors of p38MAPK and ERK kinase pathways, ERK and p38MAPK protein kinase pathways could influence the FSE-induced transcription of phase II detoxifying enzymes and transporters.
In summary, PET activation of Nrf2 signalling pathway to induce phase II detoxifying enzyme and transporter transcription may be associated with ERK and p38MAPK kinase pathways. The mechanisms are probably related to the roles of ERK and p38MAPK in directly phosphorylating Nrf2, promoting nuclear Nrf2 import and increasing its intranuclear accumulation. However, the underlying molecular mechanism remains to be studied further.
Acknowledgments
This work is supported by a grant from the Science and Technology of Shenyang (No. F15-169-4-00).
References
- Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci USA 1994; 91: 9926-9930.
- Kwak MK, Itoh K, Yamamoto M, Kensler TW. Enhanced expression of the transcription factor Nrf2 by cancer chemo-preventive agents: role of antioxidant response element-like sequences in the Nrf2 promoter. Mol Cell Biol 2002; 22: 2883-2892.
- McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a tethering mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem 2006; 281: 24756-24768.
- Shih PH, Yen GC. Differential expressions of antioxidant status in aging rats: the role of transcriptional factor Nrf2 and MAPK signalling pathway. Biogerontol 2007; 2: 71-80.
- Ma T, Liu Z. Effect of p38 MAPK-HSP27 signalling pathway in endotoxin-induced lung injury in rats. Chinese J Pathophysiol 2012; 11: 1943-1949.
- Wu L, Yu JB, Gong LR. Effect of extracellular signal-regulated kinase 1/2 on heme oxygenase-1 expression in lungs of endotoxin shock rats. Int J Anesthesiol Resuscit 2013; 3: 216-219.
- Cong ZX, Wang HD, Wang JW, Pan H, Zhou Y. Effect of MEK/ERK signalling pathway on nuclear Nrf2 accumulation in human glioblastoma U87 cells. Shandong Med J 2013; 53: 14-16.
- Kun Li, Taowen P, Shanshan H, Chuanxun L, Fei W. Study on the preparation, quantification and antitumor activity of total phenolic acids in Phyllanthus emblica. Biomed Res 2015; 26: 161-168.
- Liu L, Zhang K, Tan L, Chen YH, Cao YP. Alterations in cholesterol and ganglioside GM1 content of lipid rafts in platelets from patients with Alzheimer disease. Alzheimer Dis Assoc Disord 2015; 29: 63-69.
- Morton JF. The Emblica (Phyllanthus emblica L.). Econom Bot 1960; 14: 119-128.
- Chen ZY, Liu XM, Wu JJ, Li SF, Zou YX. Biological characteristics and nutritional components of Phyllanthus emblica L. South China Fruits 2003; 32: 71-73.
- Xu YX. Chemical constituents of Phyllanthus emblica L. and technology for extraction of total phenols. Beijing Univ Chinese Med 2009.
- Surh YJ. Cancer chemoprevention with dietary phytochemicals. Nat Rev Cancer 2003; 3: 768-780.