ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Health Science and Bio Convergence Technology: Edition-II
Particle swarm optimization based workflow scheduling for medical applications in cloud
Scientific users consider cloud platform as a popular distributed computing platform for deploying large scale medical workflow applications. These applications can be executed cost-effectively in IaaS cloud service model, as it provides scientific users with infinite pool of heterogeneous resources and pay-peruse billing model. In the proposed work, the medical science applications are mapped to the IaaS resources based on the billing scheme and the billing granularity of the cloud resources. The goal of the proposed work is to schedule medical workflow applications using discrete particle swarm optimization (DPSO) for minimizing makespan and cost. The schedule generated by the DPSO is rescheduled based on the task runtime and resource billing granularity. The proposed approach is evaluated on Epigenomic medical workflow application.
Keywords
Cloud computing, Medical workflow applications, Billing model, Billing granularity
Introduction
Medical science workflow applications consists of large number of tasks which are either complex or simple, also there exists a huge amount of data transfer between the tasks. The tasks of the workflow applications are interdependent and are normally represented using directed acyclic graphs (DAG). Cloud platform is currently considered as the cost-effective distributed computing platform for scientific workflow applications [1]. Currently, most of the cloud service providers provide the resources to the customers using different billing models and charge the usage either on hourly basis or on minute basis. So scheduling of medical science workflow applications requires the user to properly select the resources. However, since there are large number of resources which are dynamic so the workflow application scheduling to cloud resources can be efficiently addressed by using meta-heuristics.
Motivation
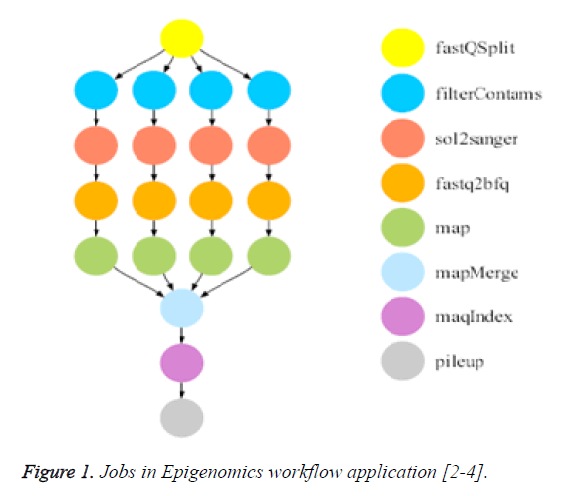
The medical workflow application used in our experiments is Epigenomics which is used at USC Epigenome center for analyzing the genome sequencing of the human body. It contains sequence of automated operations to collect short DNA segments using high-throughput gene sequencing machines and it uses MAQ software and IlluminiaSolexa genetic analyzer [2,3]. The relationship between the tasks of the Epigenomic workflow application is modelled using eight levels, with each level containing tasks that perform certain functions. The major functions in this application are splitting the data into small size chunks, reformatting chunks to a reference genome and merge the reformatted sequences into a single output map. Finally, it contains tasks that compute the sequence density for each location of interest in the reference genome [2]. Figure 1 shows the structure and tasks of the Epigenomics workflow application. This application is a data intensive workflow application and contains both large and small size tasks.
The popularity of cloud platform can be attributed to its business model which offers wide range of virtual machines (VMs) with different billing schemes and granularity. Providers such as Google Compute Engine [5] offer wide range of VMs with billing granularity in minutes and hours. For small tasks it will be better to use fine-grained billing granularity and for large tasks it is beneficial to have coarsegrained billing granularity. The advantage of coarse-grained billing model is VM overhead cost is less compared to the finegranularity and whereas fine-grained model will have less resource usage and cost. Coarse-granular model is best for big tasks and fine-grained model is good for small tasks [6]. When deploying large scale medical workflow applications, which contain both big and small tasks, it requires the scheduler to properly analyze the type, billing scheme and billing granularity of the VM and select the appropriate VM.
The influence of billing granularity on Epigenomic medical workflow application is shown in Table 1. The results show the comparison of cost and overhead values. The experiment was carried out using default Data center and VM configuration of WorkflowSim. Epigenomics workflow with different number of tasks were executed with only billing per minute resources, only Billing per Hour resources and both Billing per Hour and Billing per Minute. It can be seen from the results that there is more overhead cost associated with Billing per Minute and execution cost is more for Billing per Hour model. It also can be noticed from the results that when both kind of resources are available there is an improvement in the cost and also one more advantage is partial usage of resource is greatly reduced. Here tasks with runtime less than 5 minutes are considered as small tasks otherwise the tasks are considered as big tasks.
| Number of Tasks (Epigenomics) | Billing/Minute | Billing/Hour | Billing/Hour+Min | |||
|---|---|---|---|---|---|---|
| (Cost per minute=5, Ovehead cost=2) | ||||||
| Cost | Overhead | Cost | Overhead | Cost | Overhead | |
| 24 | 89902.9 | 147.67 | 5317380 | 2.46 | 5256388 | 4.16 |
| 46 | 209080 | 345.02 | 12422638 | 5.75 | 12345752 | 7.89 |
| 100 | 2022950 | 3361.67 | 1.21E+08 | 56.03 | 1.21E+08 | 56.82 |
| 997 | 2E+07 | 32699.6 | 1.18E+09 | 544.99 | 1.18E+09 | 556.46 |
Table 1. Executing Epigenomic workflow with different billing granularity.
In this paper, new methods are proposed to solve the medical workflow scheduling optimization problem by using discrete Particle Swarm Optimization and rescheduling the obtained solution to exploit the advantages of billing granularity of the resources. The structure of the paper is as follows:
• Related work.
• Solution for the billing model aware medical workflow scheduling using modified DPSO.
• The experimental results of the proposed algorithm.
• Conclusion and future directions.
Related work
Medical Workflow scheduling problem in cloud platform represents NP-hard optimization problem [7]. Zhou et al. propose Dyna a framework to minimize the monetary cost of executing workflow applications in cloud platform which guarantees probabilistic deadline [8]. The Dyna framework includes algorithms for dynamic configuration of resources and also authors propose hybrid configuration of instances to utilize the spot instances. Nirmala and Bhanul used a variation of traditional PSO called Catfish-PSO to schedule scientific workflow applications with the objective of reducing the makespan and execution cost [9]. The authors of Catfish-PSO based workflow do not address the billing model and billing granularity of the resources when mapping task to resources. Pietri and Sakellariou [10] propose two algorithms CSFS-Max and CSFS-Min with iterative approach to choose the resources with proper frequency. Authors have carried out performance analysis of their algorithms with three types of billing models namely Linear, Sublinear and Superliner. Bo et al. [11] propose a technique for automatically deploying and configuring the resources to schedule medical workflow application using Galaxy tool. Authors also improve the reliability of large-set data transfer in Galaxy by integrating with the features of Globus tool kit.
Huang et al. [12] propose calculation of tunable fitness function based on user’s priority for low makespan and low cost. This heuristic is extended further by using bottleneck reduction algorithm to identify and execute the task with the largest number of descendents. Wu et al. [13] propose revised discrete PSO which uses set based operations for particles position and velocity updation. Authors considers both transmission and computation cost to meet the QoS constraints for efficient scheduling of workflow applications. They consider schedule as a pair of (task, service) which learn its position from other candidate pairs which greatly reduces search space. They also analyse the performance of their method with traditional PSO and BRS (Best Resolution Scheduling).
Problem statement
In this section first a brief overview of the IaaS cloud platform is presented. Then, a solution for the medical workflow scheduling for minimizing makespan and cost in a cloud platform using discrete PSO which considers all the billing schemes and billing models of the public cloud provider is proposed.
System model
In the proposed method, we consider execution of workflows using IaaS cloud services. IaaS service providers offer different types of virtual machines to the customers. Virtual machines are created in the data centers on the physical machines using virtualization technology. Each virtual machine will be assigned number of cores, memory, storage, MIPS and bandwidth. The cost of using virtual machines in public cloud depends on the type of the virtual machine, billing scheme and billing granularity of the resources.
The objective of proposed work is to reduce the monetary cost and makespan for executing medical workflow applications in cloud environment. The monetary cost of executing medical workflow application in cloud platform represents the cumulative execution cost of all the tasks in the workflow [14,15]. Computation time or execution time of task T on virtual machine M is calculated as follows.
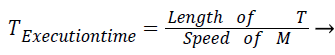 (1)
(1)
Communication cost of the task T is equal to data transfer time of task between parent and its children tasks. It is calculated as follows
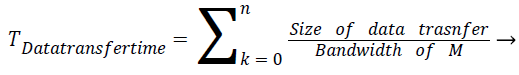 (2)
(2)
In the above equation n represents the number of parents and children of task T. From the above two equations total execution time of the task is calculated using equation 3.
 (3)
(3)
Total cost of task T is calculated based on task execution time and billing granularity of the resource using the following equation
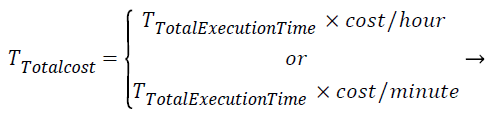 (4)
(4)
Total cost of executing a workflow W with n tasks can be calculated as follows
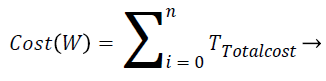 (5)
(5)
Makespan of the workflow application represents the total time required to complete workflow execution and it is computed using equation (6)
Makespan(w)=Finish_Time(TExit) → (6)
The objective of the proposed work is minimize cost and makes pan for executing medical science workflow application which is can be defined as
Minimize(Cost(W), Makespan(W)) → (7)
Traditional PSO
Traditional Particle Swarm Optimization [16] is a bio-inspired meta-heuristics which works on the principle of how the birds search for the food. Here, the candidate solutions for any problem are represented as particle and collection of particles is called swarm. The particles fly through a search space with some velocity values similar to how the birds fly in search of food and then it changes it position. The main advantage of PSO is that particles learn by self and there are no operators required for creating next generation. Each particle is associated with two values namely local best and global best. Local best represents particles best value and global value represents best value among all the particles.
DPSO with billing model for medical workflow scheduling
The challenge involved in scheduling medical workflow application in cloud is analyzing tradeoff of executing workflow task on a virtual machine and need to check large search space that is generated because of large number of resources with different resource type, billing model and billing granularity. Medical Workflow application represents class of workflow scheduling problems in cloud platform that represents NP-complete problems. The above scheduling problem represents a discrete space because either the task is scheduled on virtual machine or not scheduled. So the outcome is either 1 or 0. Hence to solve these optimization problems continuous optimization techniques such as PSO requires the conversion mechanisms because its positions are real-valued. In the proposed method, Discrete Particle Swarm Optimization which is a variant of PSO and can perform optimization in discrete domain is adopted.
Discrete particle swarm optimization
The particles of DPSO consist of binary values and it performs the optimization and the velocity represents the probability of the binary value taking value one and in our application it is probability of workflow task assigned to virtual resource. Both velocity and position take binary values in this optimization.
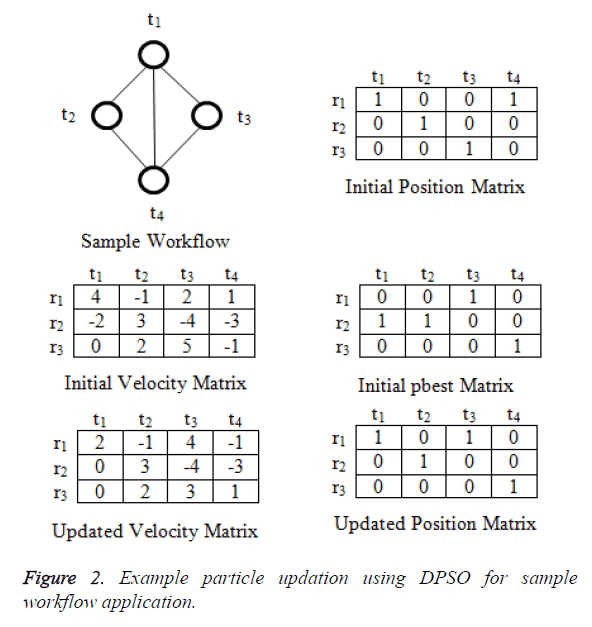
First thing in swarm based optimization technique is initialization of particles and in the given problem each particle will be matrix of size m x n with binary values. Where m represents tasks and n represents virtual machines. The values are initialized randomly. Figure 2 shows the sample workflow application and initial position for particle. Velocity of each particle is also represented using matrix with values in the range -vmax to vmax. Initial pbesti for ith particle is represented by m X n matrix. In DPSO new position of the particles and the velocity values are calculated in using the following equations [17]
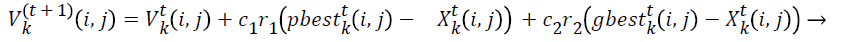 (8)
(8)
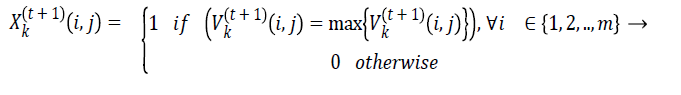 (9)
(9)
Equation (8) and (9) contain the terms of kth particle velocity and position
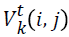 -velocity matrix element at time interval t
-velocity matrix element at time interval t
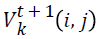 -velocity matrix element at time interval t+1
-velocity matrix element at time interval t+1
 -pbest matrix elemen at time t
-pbest matrix elemen at time t
 -gbest matrix at time t
-gbest matrix at time t
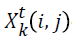 -position matrix element at time interval t.
-position matrix element at time interval t.
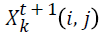 -position matrix element at time interval t+1
-position matrix element at time interval t+1
Outline of the proposed workflow scheduling is given as:
Step 1: Initialization.
Step 2: Calculation of gbest and pbest for the initial swarm
Step 3: Calculation of fitness values of each particle using equations (5) and (6)
Step 4: Calculate Position and Velocity of each particle using equation (8) and (9)
Step 5: If the fitness value is better than old gbest and pbest then Update with new gbest and pbest value.
Step 6: Repeat Steps 4 to 6 till the maximum iteration is reached.
Step 7: End
Rescheduling of tasks based on billing granularity of resources
In the proposed system after obtaining the near optimal schedule from DPSO, it is sent to the rescheduler to further improve the cost and VM overhead. For each task and VM assignment obtained, we compare the task length and the billing granularity of the VM and if the task is a small task then it is assigned to the VM with the minute granularity, otherwise the task is assigned to VM with hour granularity.
Input: Schedule S generated by DPOSO
Begin
1 For each pair of (task t,vm r) belonging to Schedule S
2 if task t is a 'small' and r.billing+granularity == 'hour'
then
{
3 for each virtual machine vmi in VM set
4 find the resource vmi with billing+granularity=='minute' assign task t to r1
}
Else if
6 task t is a 'large' and r.billing_granularity=='minute' then
{
7 for each resources vmi in the VM set
8 find the resources vmi with biling_granularity=='hour'
9 assingn t to r1
}
Repeat above steps for all the (task,resource) pairs of schedule S
End
Experimental Result Analysis and Discussion
Proposed system is implemented by conducting experiments using extension of CloudSim [18] called WorkflowSim [19]. Table 2 provides the details of different types of virtual machines and Table 3 shows the data centre configuration used in the experiment.
| S. No | Resource Type | RAM (MB) | MIPS | PE |
|---|---|---|---|---|
| 1 | Small | 2048 | 1000 | 2 |
| 2 | Medium | 4096 | 2000 | 4 |
| 3 | Large | 8192 | 4000 | 8 |
Table 2. Different types of VM.
| MIPS | 16000 |
| Number of Hosts | 20 |
| RAM | 16048 (Host Memory) |
| Storage | 1000000 (Host Storage) |
| Bandwidth | 10000 |
| Cost_per_processing | 3 |
| cost_per_mem | 0.05 |
| Cost_per_storage | 0.1 |
| Cost_per_BW | 0.1 |
Table 3. Data center configuration.
Makespan and cost comparison
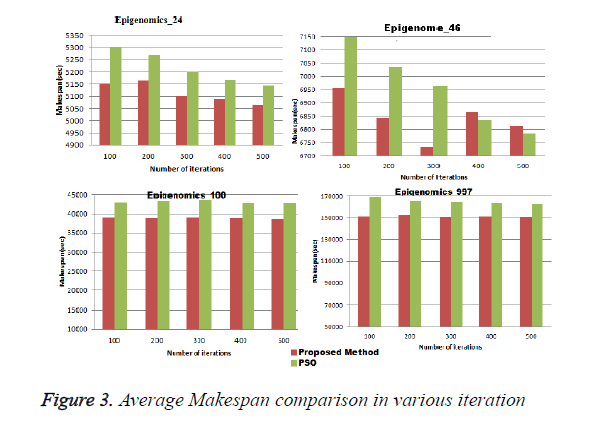
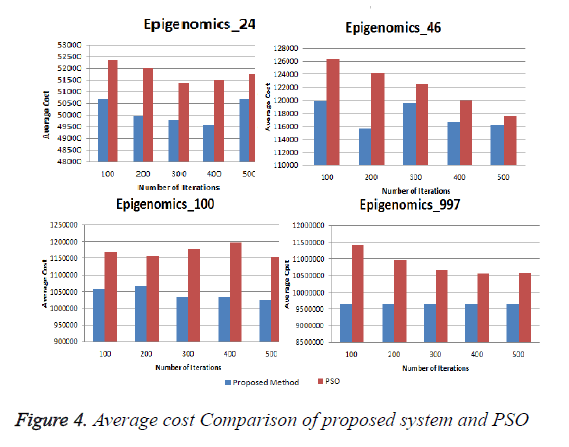
In this experiment the proposed systems was executed for different iterations for different sizes of Epigenomics workflow application and for each iteration average make span and cost was calculated. And the results were compared with the traditional PSO. Figure 3 shows the average makespan comparison ,it can be seen from the result that proposed system gives better performance compared to the traditional PSO for all the iterations. Figure 4 shows the average cost of execution for different sizes of Epigenomics workflow applications using proposed method and traditional PSO. It can be seen from the graph that the average execution cost for large size workflow applications such as Epigenomic_997 increases substantially from the proposed system.
Conclusion
Cloud computing provides lot of opportunities for executing large size medical workflow applications. Cloud service providers bill the customer’s usage by offering different billing models and billing granularity. In this work billing granularity of per hour and per minute is considered. The main contribution of the proposed work is scheduling of medical workflow application using discrete particle swarm optimization and the obtained schedule is rearranged to get the benefits of billing granularity. Initially, the performance analysis of executing small, medium and large medical workflow applications with different type of resource is carried out. Then the comparison of the average makespan and cost of executing medical workflow application using proposed method and traditional PSO is carried out. Results show that proposed DPSO algorithm with rescheduling according to billing granularity gives better performance for Epigenomic workflow application when compared with the traditional PSO.
References
- Hoffa C, Mehta G, Freeman T, Deelman E, Keahey K, Berriman B, Good J. On the use of cloud computing for scientific workflows. IEEE Fourth Int Conf In eScience 2008; 640-645.
- Gil Y, Deelman E, Ellisman M, Fahringer T, Fox G, Gannon D, Goble C, Livny M, Moreau L, Myers J. Examining the Challenges of Scientific Workflows. Computer 2007; 40: 24–32.
- Deelman E, Vahi K, Juve G, Rynge M, Callaghan S, Maechling PJ, Mayani R, Chen W, Silva RF, Livny M, Wenger K. Pegasus: a Workflow Management System for Science Automation. Future Gen Comp Sys 2015; 46: 17-35.
- https://pegasus.isi.edu
- Gonzalez JLU, Krishnan SPT. Building Your Next Big Thing with Google Cloud Platform: A Guide for Developers and Enterprise Architects. Apress 2015.
- Al-Roomi M, Al-Ebrahim S, Buqrais S, Ahmad I. Cloud computing pricing models: a survey. Int J of Grid Dist Comp 2013; 6: 93-106.
- El-Rewini H, Lewis TG. Task scheduling in parallel and distributed systems. Prentice-Hall 1994.
- Zhou AC, He B, Liu C. Monetary cost optimizations for hosting workflow-as-a-service in IaaS clouds. IEEE Trans. Cloud Comput vol 2015; 99: 1.
- Nirmala SJ, Bhanu SMS. Catfish-PSO based scheduling of scientific workflows in IaaS cloud. Computing 2016; 98: 1091-1109.
- Ilia P, Rizos S. Cost-efficient CPU Provisioning for Scientific Workflows on Clouds. GECON, 2015.
- Liu B, Madduri RK, Sotomayor B, Chard K, Lacinski L, Dave UJ, Li J, Liu C, Foster IT. Cloud-based bioinformatics workflow platform for large-scale next-generation sequencing analyses. J Biomed inform 014; 49: 119-133.
- Wu K. A tunable workflow scheduling algorithm based on particle swarm optimization for cloud computing 2014.
- Huang, J., Wu, K., Leong, L.K., Ma, S., Moh, M. A tunable workflow scheduling algorithm based on particle swarm optimization for cloud computing. Int J Soft Comput Softw Eng 2013; 351–358.
- Tirapat T, Udomkasemsub O, Li X, Achalakul T. Cost optimization for scientific workflow execution on cloud computing. In Parallel and Distributed Systems (ICPADS) Int Conf 2013; 663-668.
- Alkhanak EN, Lee SP, Khan SR. Cost-aware challenges for workflow scheduling approaches in cloud computing environments: Taxonomy and opportunities. Fut Gen Comp Sys 2015; 50: 3-21.
- Kennedy J, Eberhart R. Particle swarm optimization. IEEE Int Conf Neu Net 1995; 4: 1942–1948.
- Izakian H, Ladani BT, Abraham A, Snasel V. A discrete particle swarm optimization approach for grid job scheduling. Int J Innov Comp Inf Cont 2010; 6: 4219-4233.
- Calheiros RN, Ranjan R, Beloglazov A, Rose CAF, Buyya R. CloudSim: a toolkit for modeling and simulation of cloud computing environments and evaluation of resource provisioning algorithms. Software 2011; 41: 23-50.
- Chen W, Deelman E. Workflowsim: A toolkit for simulating scientific workflows in distributed environments. IEEE In E-Science 8th Int Conf 2012; 1-8.