ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2014) Volume 25, Issue 4
Noninvasive prenatal analysis by Multiplex fluorescent PCR of maternal plasma DNA.
1Department of Clinical Laboratory, Renmin Hospital of Wuhan University Wuhan, 430060, P.R. of China
2Department of pediatrics, Hubei Maternal and Child Health Hospital, Wuhan, 430070, P.R. of China
- *Corresponding Author:
- Tang Dong-ling
Department of Clinical Laboratory
Renmin Hospital of Wuhan University
Wuhan, 430060, P.R. of China
Accepted date: June 21 2014
The value of circulating fetal free DNA in maternal plasma for clinical applications was evaluated. The DNA template was extracted from 32 pregnant women and the whole blood samples isolated from the stated biological fathers were used to detect genotype. Using the nested polymerase chain reaction (PCR), the 130bp SRY gene-specific sequence and the 261bp ATL1 gene-specific sequence were amplified simultaneously. The results were confirmed after delivery. Multiplex PCR amplification at sixteen polymorphic short tandem repeat (STR) loci was also used to detect the fetal DNA in maternal plasma. The SRY genespecific nested PCR product was detected in 19 plasma samples obtained from pregnant women, deducing they bear the male fetus and the remaining pregnant women bear female. When compared with the birth outcome, one sample was false positive. The coincidence was 96.88%. Multiplex fluorescent PCR with 16 polymorphic short tandem repeats revealed the presence of fetal DNA in all cases. Circulating fetal DNA analysis can be used as a possible alternative tool in routine laboratory prenatal diagnosis in the near future. This assay provides a sensitive, accurate and efficient method for noninvasive prenatal genetic diagnosis.
Keywords
Fetal DNA, Maternal plasma, Short tandem repeat, Non-invasive prenatal diagnosi
Introduction
Prenatal diagnosis of fetal genetic conditions is a standard part of modern obstetric care. Many of the current methods rely on invasive methods and are associated with an inherent risk of fetal loss. Consequently, there has been a long-term goal for development of non-invasive prenatal diagnostic methods [1-5].
Recently, much interest has been focused on the biology and diagnostic applications of nucleic acids that are present in the plasma and serum of human [6-10]. In particular, fetal DNA has been found to exist in maternal plasma [11]. This discovery has opened up new possibilities for the non-invasive prenatal diagnostic approaches based simply on the analysis of a maternal blood sample. However, a technical challenge experienced by many researchers in the field relates to the ability to discriminate fetal DNA from the coexisting background of maternal DNA in maternal plasma. During pregnancy, fetal DNA amounts to 3–6% of the total DNA in maternal plasma [12]. Hence, the diagnostic reliability of fetal DNA analysis in maternal plasma depends on the sensitivity and specificity of the analytical system for the detection of fetal-specific markers. The first marker that was developed for fetal DNA detection in maternal plasma was the Y chromosome. However, it is only applicable to pregnancies involving male fetuses. Applications of plasma DNA polymerase chain reaction (PCR) to pregnancies involving female fetuses require the development of PCR systems that are capable of detecting autosomal polymorphic loci. Amplification of highly polymorphic short tandem repeat (STR) markers outside the Y chromosome should be suitable for this purpose [13].
The goal of this study was to probe the feasibility and scientific value of fetal DNA in maternal plasma for noninvasive prenatal diagnosis in two ways. One way is to develop an efficient nested PCR-based system for detection of Y-chromosome-specific sequence (SRY) gene in fetal DNA; The other is to use PCR amplification of sixteen different polymorphic STR, we could demonstrate that fetal DNA can be detected by the presence of paternally inherited fetal-specific alleles in maternal plasma samples by two ways. These data may improve our understanding of the fetal DNA characteristic in maternal blood and thus give some indications for assessing the potential of circulating fetal DNA analysis as a possible alternative tool in routine laboratory prenatal diagnosis.
Materials and methods
Subjects
A network of three clinical sites was established to gather specimens. Clinical sites received institutional review board approval before patient enrollment. 32 pregnant women aged from 20 to 35 years, who were between the 8th and the 37th week of a singleton gestations and who did not have serious anemia (Hb>90 g/l) or any other serious complication of pregnancy were included, the stated biological fathers also participated in the study. Written informed consent was obtained before participation. All test results were compared with amniocentesis or newborn reports obtained from the clinical sites.
Maternal blood (5 ml) was collected into one tube containing ethylene diamine tetraacetic acid (EDTA) in each case. Specimens were picked up the same day at regional clinical sites or shipped by commercial carrier for overnight delivery. Blood was stored at 4°C until processed. The samples were shipped to our central laboratory for analysis. Laboratory personnel were masked as to the identity of the samples with a numerical coding system. Control blood samples were also taken from healthy men and nonpregnant women, respectively.
Plasma DNA preparation
Maternal blood samples were centrifuged at 2000g for 15 minute, and the plasma was carefully removed from the collection tubes and transferred into plain polypropylene tubes. The plasma samples were then re-centrifuged at 2000g for 10 minute, and the supernatants were collected into fresh tubes. The samples were stored at -20°C until further processing.
DNA extraction
DNA was extracted from plasma samples using the QIAamp DNA Blood Midi Kit (Qiagen) with the blood and body fluid protocol supplied by the manufacturer. We used plasma sample (2 ml) per column for DNA extraction. Genomic DNA from peripheral leukocytes and amniotic fluid was performed by standard methods in our laboratory [14].
PCR analysis
Fetal sex was detected in 32 pregnant women by identifying the SRY gene in maternal plasma using nested PCR analysis. Two rounds of PCR were carried out to amplify the fragment of the SRY gene. The first PCR analysis was performed in a total volume of 50μl. The PCR mixture contained 5-10μl extracted DNA, 5μl PCR buffer, 200μmol/l of dNTPs, 20pmol “external” SRYspecific oligonucleotide (Y1 5-GTG TCC TCT CGT TTT GTG AC-3; Y2 5-GTA ATC ATC GCT GTT GAA TAC-3) and 0.5U Taq DNA polymerase (Promega, USA). The PCR conditions were preheating at 95°C for 5 min, then 30 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 45 s, and then 72°C for 10 min. The size of the PCR fragment was 369 bp.
The second round was also performed in a total volume of 50μl. The PCR mixture contained 5–10μl of the product of the first PCR round, 5μl PCR buffer, 200μmol/l of dNTPs, 20pmol “internal” SRY-specific oligonucleotide (Y3 5-TGG CGA TTA AGT CAA ATT CGC-3; Y4 5-CCT AGT ACC CTG ACA ATG TAT-3) and 0.5U Taq DNA polymerase. PCR conditions were preheating at 95°C for 5 min, then 30 cycles of 94°C for 30 s, 57°C for 30 s, 72°C for 1 min, and then 72°C for 10 min. The final PCR fragment was 130 bp. The control PCR for a fragment of ATL1 was carried out as standard assay in a total volume of 50μl. The ATL1 is the sequence in the FMR1 gene located on the long arm of the X chromosome. Oligonucleotide primers (X1 5-TCG CCT TTC TCA AAT TCC AAG-3; X2 5-CAT CCA GAG CGT CCC TGG CTT-3) were designed to amplify it. PCR conditions were preheating at 95°C for 5 min, then 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min, and then 72°C for 10 min. The size of the PCR product was 261bp. The PCR products were separated by 8% polyacrylamide electrophoresis and visualized by exposure to ultraviolet light after ethidium bromide staining. Identifications of the X-specific and Y-specific sequences in amniotic fluid and maternal genomic DNA specimens as well as positive and negative control DNA were performed in the same manner but with about 100ng purified DNA as template. Blank control was also performed throughout the PCR process using water.
The AmpFlSTR Profiler Plus PCR Amplification Kit was utilized to amplify the following tetrameric STR loci: D3S1358, D5S818, D8S1179, D13S317, D18S51, D21S11, FGA, VWA, D19S433, TPOX, TH01, D7S820, D16S539, Amelogenin, CSF1PO and D2S1338. Amplifications were conducted according to manufacturer recommendations using TC 9600 instrument (PE, USA). Analysis of the fluorescently labeled amplified fragments was resolved on ABI Prism 310 Genetic Analyzer (ABI, USA) by Genescan software. PCR amplification was performed in a final volume of 25 μl and contained PCR reaction mix 10μl, Taq Gold DNA Polymerase 5U/μl) 0.5μl, Identifier primer set 4.5μl. We used 10 μl template extracted from plasma sample for amplification. Initial incubation at 95°C for 11 min was followed by denaturation (94°C for 1min), annealing (59°C for 1min) and extension (72°C for 1min) for 32 cycles and at the end there was an incubation at 60 °C for 60 min. PCR products were mixed with 20μl of formamide (Sigma- Aldrich, St. Louis, MO, USA) and 1μl of Prism Genescan-500 TAMRA size standard (PE Applied Biosystems, Warrington, Great Britain). The mixture was denatured at 95 °C for 3 min and cooled to 4 °C for 5 min. Electrophoresis was performed on ABI 310 Genetic Analyser by using POP gel (PE Applied Biosystems, Fosters City, CA, USA). The results were analyzed with Genescan Analysis software.
Anticontamination measures
Great care was taken to prevent PCR contamination. Aerosol-resistant pipette tips were used for all liquids. All procedures including blood specimen preparation, DNA extraction and PCR amplification were performed by female members in a separate area. Multiple water blanks were included as controls in every analysis. All the samples were analyzed in duplicate.
Results
SRY gene analysis of plasma DNA
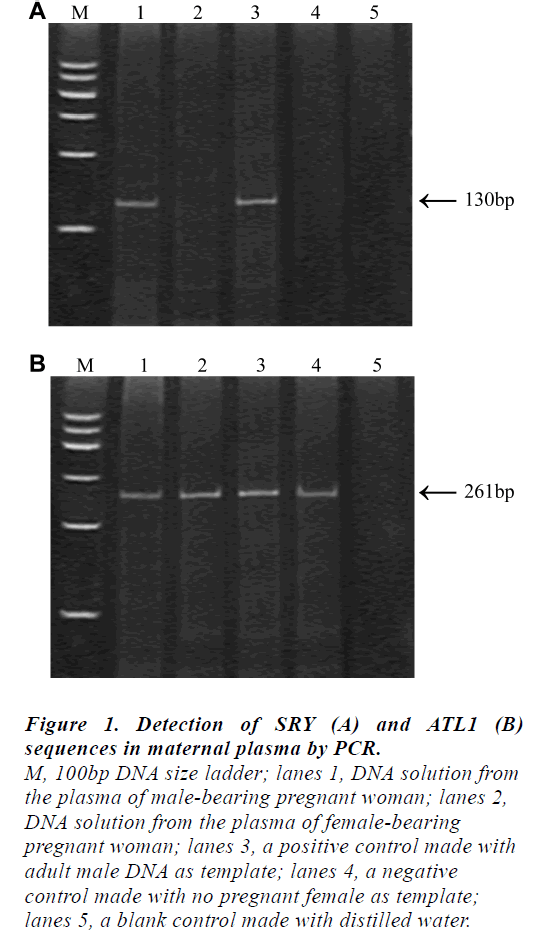
The results obtained by the non-invasive plasma DNA analysis of the ATL1 and SRY sequences in representative individuals were shown in Fig 1. The presence of the 130 bp fragments in the plasma sample would represent the fetal origin (lane1) while the 261 bp ATL1-specific fragments could be observed in all cases.
Figure 1: Detection of SRY (A) and ATL1 (B)
sequences in maternal plasma by PCR.
M, 100bp DNA size ladder; lanes 1, DNA solution from
the plasma of male-bearing pregnant woman; lanes 2,
DNA solution from the plasma of female-bearing
pregnant woman; lanes 3, a positive control made with
adult male DNA as template; lanes 4, a negative
control made with no pregnant female as template;
lanes 5, a blank control made with distilled water
It is noteworthy that the 261 bp ALT1-specific fragment could be assigned as an internal control for the PCR amplification in this system; without it, fetal sex prediction would be unreliable. We have applied this system for the prenatal fetal sex prediction of 32 pregnant women at various gestational ages ranging from 8 to 37 weeks. Among 32 samples, the SRY gene amplified products were detected in 19 samples, which were diagnosed as male fetus while no SRY gene amplified product was detected in 13 samples, which were diagnosed as female fetus. The results were compared with those obtained from routine analysis of amniotic fluid specimens and the fetal sex outcome at birth. No false negative sample and one case of false positive samples (case12) were found, who previously had a spontaneous abortion. The total concordance rate was 96.88% (31/32).
STR analysis of plasma DNA
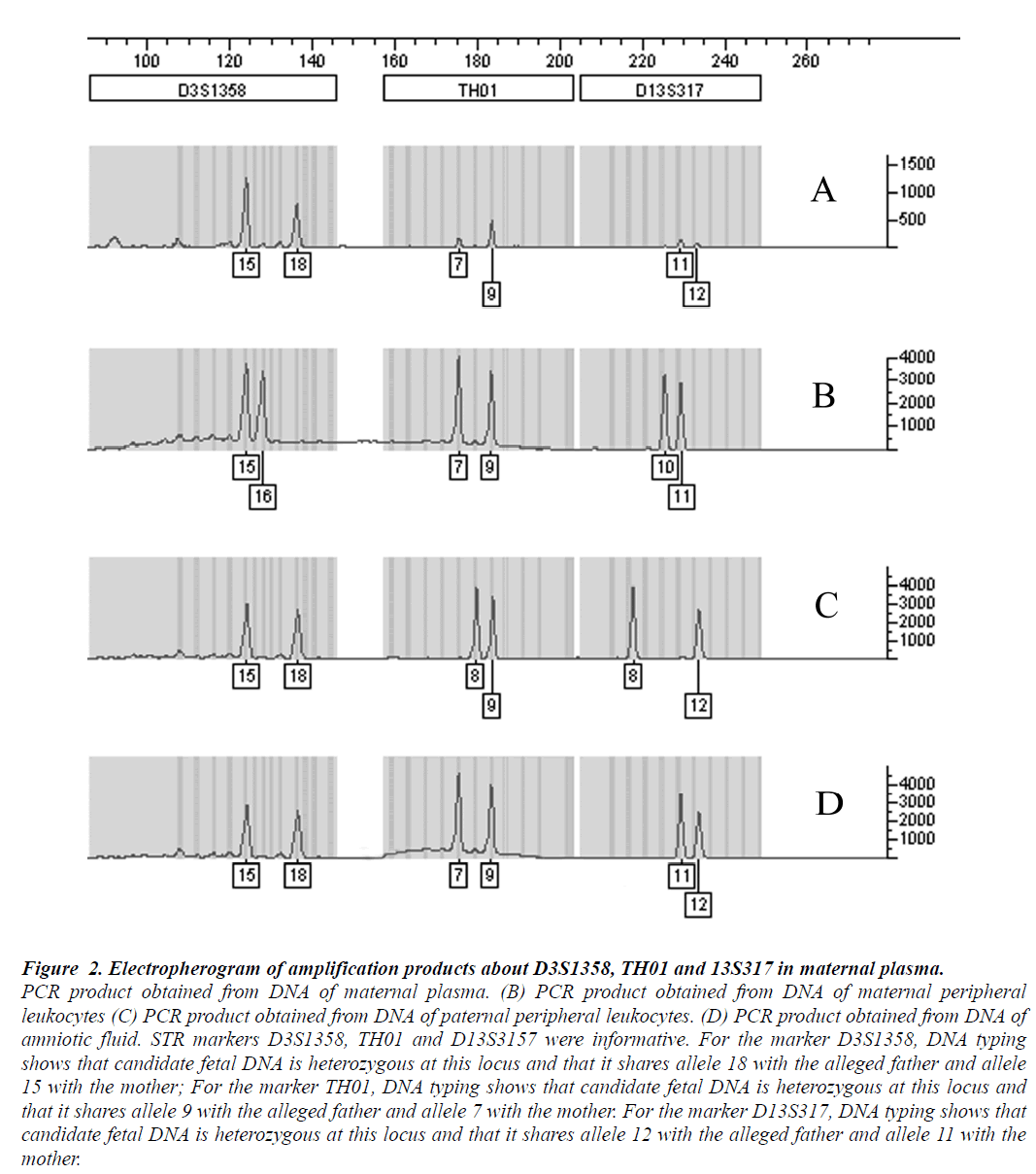
Multiplex fluorescent PCR with polymorphic short tandem repeats, used to analyze maternal plasma samples collected before delivery, revealed the presence of fetal DNA in all cases. An example of the alleles demonstrated in maternal plasma was shown in Fig 2, For the marker D3S1358, the allele 18 was not found in the analysis of the maternal whole blood DNA, corresponding to the paternally inherited allele of the fetus. For the marker TH01, the allele 9 corresponded to the paternally inherited allele of the fetus and the allele 12 indicating the paternally inherited fetal allele from the marker D13S3157. Candidate fetal DNA sharing one with the maternal WBCs, but containing another allele that is clearly different, suggests that it is indeed fetal in origin.
Among the 32 pairs of samples, all the cases were informative with at least one STR marker. The earliest gestational age at which fetal DNA was detected was 8 weeks. These results suggested that STR analysis of maternal plasma could potentially be used for early noninvasive prenatal diagnosis.
Figure 2: Electropherogram of amplification products about D3S1358, TH01 and 13S317 in maternal plasma.
PCR product obtained from DNA of maternal plasma. (B) PCR product obtained from DNA of maternal peripheral
leukocytes (C) PCR product obtained from DNA of paternal peripheral leukocytes. (D) PCR product obtained from DNA of
amniotic fluid. STR markers D3S1358, TH01 and D13S3157 were informative. For the marker D3S1358, DNA typing
shows that candidate fetal DNA is heterozygous at this locus and that it shares allele 18 with the alleged father and allele
15 with the mother; For the marker TH01, DNA typing shows that candidate fetal DNA is heterozygous at this locus and
that it shares allele 9 with the alleged father and allele 7 with the mother. For the marker D13S317, DNA typing shows that
candidate fetal DNA is heterozygous at this locus and that it shares allele 12 with the alleged father and allele 11 with the
Discussion
Recent discovery of the presence of circulating cell free fetal DNA in maternal plasma has raised many unanswered biological questions and opened up numerous diagnostic applications [15-19]. In our study, the first purpose was to detect fetal DNA to develop a non-invasive prenatal diagnosis technique, and fetal sex detection was considered as a successful assay. Though the nested PCR method cannot be comparable to the real-time quantitative PCR approach [20], it was indicated that the method can provide a more practical methodology in clinical researches and diagnosis with acceptable sensitivity and specificity, especially in some developing countries. Unlike other described PCR methods for prenatal fetal sex determinations in maternal plasma [21], the incorporation of the Xspecific ATL1 gene amplification as an internal control of the nested PCR system herein described could greatly improve the reliability of the fetal sex prediction. As shown in Fig1, we were able to identify the fetal sex using nested PCR analysis of 32 maternal plasma samples during 8–37 weeks of gestation, assessed in blinded experiment as compared to the conventional amniotic fluid analysis and the birth outcome.
The reliability of nested PCR assay used was 96.88% .We obtained false positive result in one sample; case12 previously had a spontaneous abortion. For the risk of false positive result in this study, we could not ignore the possible effects of fetal cells from former pregnancies. The fetal DNA we detected in maternal plasma derived from the continuous lysis of residual fetal cells in the light of the well-known presence of entire fetal cells in maternal circulation during the same period. It is unlikely that free DNA circulates for years in the blood without being degraded by plasma nucleases or other organ systems. Another risk is the formation of non-specific products, during the experiments, male DNA contamination can be magnified by nested PCR and conceals the real results, especially when the primer specificity or the extension temperature are low, or the extending probability of mismatched nucleotides were largely decreased due to the substitution of polymerase with high fidelity for common Taq polymerase. All this may lead to the false positive result we obtained in the fetal DNA detections. Therefore, it is wise to search for optimum conditions and carry out the experiment strictly according to the instructions so as to evade false results as much as possible.
Although the method did not completely agree with the birth outcome, and need improving, it was still a useful tool for non-invasive prenatal diagnosis because of its high sensitivity, better reliability and simple manipulation compared with the common amplification of specific sequence in Y chromosome. The nested PCR approach described here would be more practical in any laboratory where a conventional PCR is available. It may be useful in place of invasive prenatal diagnostic methods such as amniocentesis and chorionic villi sampling in cases of screening X-linked recessive inheritance. However, it cannot be used for the 50% of pregnancies involving female fetuses, thus hindering the translation of these promising observations into the realms of clinical use. STR typing, the most powerful technique for individualization of biological stains, represent a major advantage over other DNA detection systems that use fetus derived Y sequences.
Our subsequent results demonstrate the feasibility of detection of fetal DNA in maternal plasma independent of gender with 16 highly polymorphic STR markers and fluorescent multiplex PCR. Amplification of a STR marker will result in two PCR products in most normal samples, each of which has been inherited from one parent. As shown in Fig 2, candidate fetal DNA having one allele shared with the maternal peripheral leukocytes while the sond allele is clearly different from the maternal DNA but shared with the paternal peripheral leukocytes suggests it is indeed fetal in origin.
In our study, the earliest gestational age at which fetal DNA was detected was 8 weeks. This suggests that STR analysis of maternal plasma can be used for early non-invasive prenatal diagnosis. However, the sensitivity of detecting the minority fetal allele was lower than in previous reports that utilized fetus-specific amplification schemes [12,22]. The lower level of sensitivity may be due to the nonselective nature of PCR amplification of STR (both the majority maternal alleles and the minority fetal alleles were amplified together). Under these conditions, the excess of the background sequences could out-compete the rare target sequences for amplification, but our proposed technique was sensitive enough to detect fetal-specific alleles in all pairs because of high concentration of fetal DNA present in maternal plasma. With the development of polymorphic STR markers, we believe that molecular analysis of fetal DNA in maternal plasma can be used for the non- invasive detection of fetal-derived paternally inherited aneuploidy. This report extended the clinical utility of maternal plasma DNA amplifications and made a prospective analysis of pregnancy complications feasible independent of fetal gender.
The current prenatal diagnosis approaches have relied on the use of fetal material which has been obtained through invasive means, such as amniocentesis, which carries with it a certain risk to the fetus, and of further sensitizing the mother against fetal antigens. Thus, such diagnostic tests would be limited to a proportion of cases. Fetal DNA detection from maternal plasma is a promising technique but that more experience in this field is needed. Introduction of new methods, such as fully automated systems for nucleic acid purification, together with rapid PCR systems and non-PCR amplification strategies [23], will further facilitate high throughput molecular analysis. We are in urgent need of standardization of techniques, careful evaluation and data analysis. Easy, cheap and faster techniques in future can make fetal DNA identification and quantification a routine biochemical laboratory investigation. It also eliminate the conventional methods of tissue biopsies, CT scan and invasive prenatal diagnostic tests like chorionic villus sampling (CVS) and amniocentesis. In short, Fetal DNA detection from maternal circulation is very challenging but has enormous utility if adequately managed.
Acknowledgements
This work was supported by a grant 81102141 from National Science Foundation of China
References
- Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, Leung TY, Foo CH, Xie B, Tsui NB, Lun FM. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci USA 2008; 105 (51): 20458-20463.
- Cui YX, Xia XY, Bu Y, Zhou GH, Yang B, Lu HY, Shi YC, Pan LJ, Huang YF, Li XJ. Rapid Molecular Prenatal Diagnosis of Spondyloepiphyseal Dysplasia Congenita by PCR-SSP Assay. Genet Test 2008; 12(4): 533-536.
- Sekizawa A, Purwosunu Y, Matsuoka R, Koide K, Okazaki S, Farina A, et al: Recent advances in noninvasive prenatal DNA diagnosis through analysis of maternal blood. J Obstet Gynaecol Res 2007; 33(6): 747-764.
- Findlay I, Taylor A, Quirke P, Frazier R, Urquhart A: DNA fingerprinting from single cells. Nature 1997; 389: 555-556.
- Erdal Eren, Esra Deniz Papatya Cakir, Ozlem Bostan, Halil Saglam, Omer Tarim Evaluation of the endocrine functions in pediatric patients with cyanotic congenital heart disease. Biomed Res- India 2013; 24(2): 211-215
- Wright CF, Burton H. The use of cell-free fetal nucleic acids in maternal blood for non-invasive prenatal diagnosis. Hum Reprod Update 2009; 15(1): 139-151.
- Tong YK, Chiu RW, Leung TY, Ding C, Lau TK, Leung TN, Lo YM. Detection of restriction enzyme-digested target DNA by PCR amplification using a stem-loop primer: application to the detection of hypomethylated fetal DNA in maternal plasma. Clin Chem 2007; 53(11): 1906-1914.
- Tong YK, Lo YM: Diagnostic developments involving cell-free (circulating) nucleic acids. Clin Chim Acta 2006; 363: 187-196.
- Lo YM: Circulating nucleic acids in plasma and serum: an overview. Ann N Y Acad Sci 2001; 945: 1-7.
- Galbiati S, Smid M, Gambini D, Ferrari A, Restagno G, Viora E, Campogrande M, Bastonero S, Pagliano M, Calza S. Fetal DNA detection in maternal plasma throughout gestation. Hum Genet 2005;117 (2-3): 243-248.
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, Wainscoat JS. Presence of fetal DNA in maternal plasma and serum. Lancet 1997; 350 (9076): 485-487.
- Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis.Am J Hum Genet 1998; 62(4): 768-775.
- Cha DH, Khosrotehrani K, Bianchi DW, Johnson KL. The utility of an erythroblast scoring system and genderindependent short tandem repeat (STR) analysis for the detection of aneuploid fetal cells in maternal blood. Prenat Diagn 2005; 25(7): 586-591.
- Siriratmanawong N, Fucharoen G, Sanchaisuriya K, Ratanasiri T, Fucharoen S. Simultaneous PCR detection of beta - thalassemia and alpha - thalassemia 1 (SEA type) in prenatal diagnosis of complex thalassemia syndrome. Clin Biochem 2001; 34 (5): 377-380.
- Wataganara T, Bianchi DW: Fetal cell-free nucleic acids in the maternal circulation: new clinical applications. Ann N Y Acad Sci 2004; 1022: 90-99.
- Simpson JL, Bischoff F: Cell-free fetal DNA in maternal blood: evolving clinical applications. JAMA 2004; 291: 1135-1137.
- Ding C, Chiu RW, Lau TK, Leung TN, Chan LC, Chan AY, Charoenkwan P, Ng IS, Law HY, Ma ES. MS analysis of single-nucleotide differences in circulating nucleic acids: Application to noninvasive prenatal diagnosis. Proc Natl Acad Sci 2004; 101: 10762-10767.
- Tsujie T, Takemura M, Kimura T, Shimoya K, Tsutsui T, Ogita K, Ozaki M, Murata Y. Rapid detection of trisomy 21 by gene dosage analysis using quantitative real-time polymerase chain reaction. J Obstet Gynaecol Res 2006; 32: 368-372
- Leung TY, Chan LW, Law LW, Sahota DS, Fung TY, Leung TN, Lau TK. First trimester combined screening for Trisomy 21 in Hong Kong: outcome of the first 10,000 cases. J Matern Fetal Neonatal Med 2008; 13: 1-5.
- Zimmermann B, El-Sheikhah A, Nicolaides K, Holzgreve W, Hahn S. Optimized real-time quantitative PCR measurement of male fetal DNA in maternal plasma. Clin Chem 2005; 51: 1598-1604.
- Honda H, Miharu N, Ohashi Y, Ohama K. Successful diagnosis of fetal gender using conventional PCR analysis of maternal serum. Clin Chem 2001; 47: 41-46.
- Erdal Eren, Esra Deniz Papatya Cakir, Ozlem Bostan, Halil Saglam, Omer Tarim, Lo YM, Hjelm NM, Fidler C, Sargent IL, Murphy MF, Chamberlain PF, Poon PM, Redman CW, Wainscoat JS. Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. N Engl J Med 1998; 339: 1734-1738.
- Godinho AB, Cardoso E, Melo MA, Gonçalves M, Da Graca LM. Ultrasonographic diagnosis of fetal ovarian cysts: five cases in five years. J Matern Fetal Neonatal Med 2008; 21 (12): 875-879.