ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 10
Nanoparticle-based MRI of metastatic human hepatocellular carcinoma: Targeting survivin gene with siRNA in an animal model
Rui Jiao, Gonghua Dai, Wei Chen and Peijun Wang*
Department of Radiology, Tongji Hospital of Tongji University, Shanghai 200065, PR China
- *Corresponding Author:
- Peijun Wang
Department of Radiology
Tongji Hospital of Tongji University, PR China
Accepted date: March 04, 2017
Objective: This study was designed to investigate the value of nano-MRI compared with the routine gadolinium diethylene triaminepenta acetate contrast MRI of metastatic hepatocellular carcinoma.
Methods: Livers of healthy BALB/c nude mice were implanted with cells of hepatocellular carcinoma - HepG2 and SMMC77219, respectively. Small interfering RNA targeting was accomplished against one of the three hepatocellular carcinoma-survivin gene sequences. Nanometer grade particles especially the superparamagnetic iron oxide nanoparticle superparamagnetic iron oxide nanoparticles Resovist was complexed with polylysine and siRNA designed specifically to create an enhanced contrast agent with superparamagnetic iron oxide nanoparticles. Both hepatocellular carcinoma -HepG2 and SMMC7721 groups were scanned at 1.5 T MRI without any contrast MRI, gadolinium diethylene triaminepenta acetate contrast medium, and the specific enhanced contrast agent with superparamagnetic iron oxide nanoparticles by injecting the contrast media via tail vein. All mice were scanned with T2 weighted imaging 24 h post-injection, to observe the effect of the specific contrast medium with superparamagnetic iron oxide nanoparticles. Subsequently, all the mice were sacrificed to extract the livers.
Results: Tumor detection rate 24 h post-injection, corresponding to the specific contrast agent with superparamagnetic iron oxide nanoparticles showed a significant difference compared with the gadolinium diethylene triaminepenta acetate contrast agent. The livers still showed remarkably low signal change within the signal enhanced region of T2WI compared with instant imaging using the specific SPIO.
Conclusion: Nanometer grade particles can be stably transfected into tumor cells to facilitate detection of human hepatocellular carcinoma xenografts in nude mice using small interfering RNA. The strong paramagnetic effect observed with the high valence iron in T2WI improves the detection rate of micronodular (<5 mm) HCC with MRI.
Keywords
Molecular imaging, Magnetic resonance imaging, Hepatocellular carcinoma, Survivin, Superparamagnetic iron oxide nanoparticles, Nude mice, Gadolinium diethylene triaminepenta acetate.
Introduction
Advances in molecular imaging and nanotechnology have improved the detection of high-expression oncogenes for early diagnosis of malignant tumors. Single-stranded ribonucleic acid (RNA) specifically inhibits the expression of target genes in situ mediated by RNA interference. Compared with gene knockout, RNA interference inhibits the expression of specific target genes effectively and more conveniently. As a molecular diagnostic imaging tool, it RNA interference enables detection of increased expression of malignant tumor-specific genes. It, therefore, plays an important role in the early clinical diagnosis and treatment of malignant tumors.
This study aims to leverage the components of endocytotic pathway in the design of small interfering RNA (siRNA) of survivin genes in human primary liver cancer. Using a specific molecular imaging contrast agent obtained by combining RNA interference with the super paramagnetic iron oxide nanoparticle (SPION) Resovist we compared the imaging results with conventional gadolinium diethylene triaminepenta acetate (Gd-DTPA) contrast media, and investigated the role of nano- magnetic resonance imaging (MRI) in screening for tumors orthotopically implanted in the liver of nude mice. We provide test data for clinical application of molecular imaging. Transfection of siRNA into the cytoplasm of target cells is mediated by phagotrophy. A large number of studies investigating in vitro RNA transfection suggest that binding of cationic liposomes with antibodies and ligands/receptors, can significantly improve the efficiency of siRNA transfection resulting in significant RNA interference effects [1].
The contrast agent Resovist is a dextran-coated SPION, mainly phagocytosed by reticuloendothelial system after intravenous injection. Clinically, it is used as a tissue-specific MRI contrast agent, for the diagnosis of liver tumors. Cellular uptake of Resovist causes local magnetic field inhomogeneity. T2 relaxation, the decay of transverse magnetization caused by a combination of spin-spin relaxation and magnetic field inhomogeneity, significantly reduces the signal strength of the T2 weighted images (T2WI). Due to the low signal strength, it has good negative contrast. A minute amount of contrast agent is sufficient to achieve a significantly lower signal. Compared with the conventional metal gadolinium-containing contrast agents, Resovist has higher sensitivity. Li et al. used aminosilane Fe3O4 nanoparticles to label lung adenocarcinoma transplanted into nude mice for MRI. The findings indicate that super paramagnetic iron oxide particles are endocytosed by cells for magnetic resonance imaging after combining with positively-charged transfection agents [2].
Resovist has been used clinically for MR imaging. With a particle size of 50~80 nm, it manifests stable physical and chemical properties, without any toxicity. Due to lower molecular size, it enters cells easily. The SPIO has high MR sensitivity, requiring only a trace amount to generate low signals in MR T2WI or T2* images. The dextran-coated molecule carries negative charge. Therefore, a polylysine (PLL) is used as a transfection agent carrying a large amount of positive charge on the surface to effectively bind Resovist particles by electrostatic interaction. Resovist cannot enter tumor cells and is mostly phagocytosed by reticuloendothelial cells, such as Kupffer cells (KC) in the hepatic sinusoid region. Since there are no reticuloendothelial cells in tumor, SPION cannot be phagocytosed. It manifests relatively higher signal in T2WI, and can be distinguished from normal liver tissue [3]. This negative contrast shows no specificity for tumor cells, so abnormal tissues without phagocytes have similar MR imaging. Therefore, in this study, we combined Resovist and siRNA to design a specific MR contrast agent.
Materials and Methods
Experimental animals
Healthy BALB/c nude mice, including 25 males and 25 females, aged 3-4 weeks, each weighing 20 ± 5 g, were provided by Shanghai SLAC Laboratory Animal Co., Ltd., and Shanghai Institute of Materia Medica Animal Center and fed in SPF conditions in Experimental Animal Center, Tongji Hospital, Tongji University. All animals were housed in microisolator cages with free access to food and water according to the Guide for the Care and Use of Laboratory Animals. In particular, any effort was put to avoid unnecessary pain of the animals. The whole study was approved by the Institutional Animal Care Committee at Shanghai Institute of Materia Medica Animal Center.
Establishment of animal models
All the experimental animals were randomly divided into three groups: 20 in the HepG2 group, 20 in the SMMC7721 group and 10 in the control group, comprising males and females in 1:1 ratio.
In the SPF-grade animal room, the nude mice in the test groups were intraperitoneally anesthetized with 3% sodium pentobarbital on a clean surgical platform. The left lobe of the liver was exposed to direct vision after laparotomy, horizontally punctured with a fine needle and injected with human hepatocellular carcinoma HepG2 and SMMC7721 cell suspension 0.05~0.1 ml/mouse, respectively. We implanted approximately 0.75~1.5 × 106 cells, and sutured the abdomen layer by layer. The animals were fed continuously in the SPFgrade animal room and removed randomly for MRI at periodic intervals.
MRI parameters
The MRI equipment included Marconi Company’s Edge Eclipse 1.5 T superconducting magnetic resonance scanner, with a facet joint surface coil. The nude mice were intraperitoneally anesthetized with 3% sodium pentobarbital and attached to the examination plate in the prone position, with the back facing the facet joint surface coil.
Scan sequence parameters included: unenhanced axial and coronal T1WI, T2WI, T2*; super paramagnetic iron oxide contrast agent-enhanced axial and coronal T2WI, T2*; and Gd- DTPA-enhanced axial and coronal T1WI. T1WI used FSE sequence, i.e., TR 724 ms, TE 16 ms, FOV 10 mm, and thickness 3 mm. T2WI used FSE sequence, i.e., TR 4241 ms, TE 99.2 ms, FOV 10 mm, and thickness 3 mm. T2*WI used GRE sequence, i.e.,. TR 400 ms, TE 17.9 ms, FLIP 20, FOV 10 mm, thickness 3 mm.
Preparation of super paramagnetic iron oxide contrast agent
The lyophilized powder comprising Resovist, polylysine (PLL) and siRNA was diluted with double-distilled water to Resovist 50 μg/ml, PLL 1.5 μg/ml and siRNA 10 pmol/μl, shaken evenly with a high-speed shaker for 30 min, and saved in a 5°C refrigerator for later use.
MR molecular imaging of human primary liver cancer
The nude mice were divided into HepG2 and SMMC7721 groups. The mice in the HepG2 group were inoculated with HepG2. At intervals of 1 w, 3 w, 4 w, 5 w and 6 w, 4 mice in the test group and 1 mouse in the control group were randomly subjected to 1.5T MRI. The mice in the SMMC7721 group were inoculated with SMMC7721. At periodic intervals of 1 w, 2 w, 3 w, 4 w and 5 w, 4 mice in the test group and 1 mouse in the control group were randomly selected for 1.5T MRI.
MRI sequence included unenhanced abdominal axial and coronal T1WI and T2WI scans. We then performed tail vein injection combined with survivin siRNA super paramagnetic iron oxide contrast agent for enhanced MRI axial and coronal T2WI and T2*. Each mouse was injected with the specific contrast agent 0.3 ml, containing survivin siRNA 40 μg, SPION 50 μg/ml, followed 24 h later by abdominal axial and coronal T1WI, T2WI and T2* scans. We then performed tail vein injection using 1:10 Gd-DTPA contrast agent 0.1 ml, for axial and coronal T1WI.
Specimen preparation
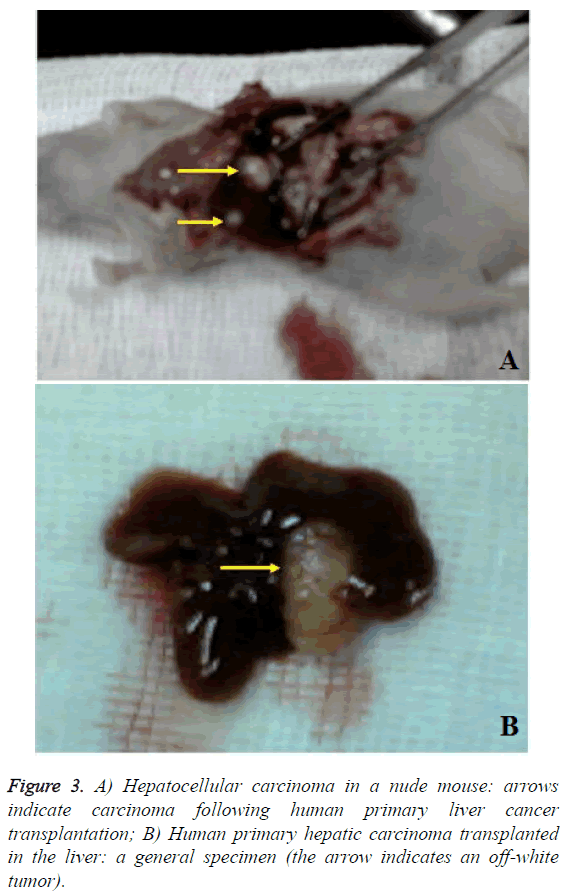
After the last MRI, the animals in the test and the control group were killed by cervical dislocation. The livers were quickly extracted and fixed with 4% paraformaldehyde solution. After conventional gradient alcohol dehydration, chloroform transparentization and embedding in paraffin, sections were obtained, followed by H&E staining and Prussian blue staining. The cell morphology and distribution of iron particlelabeled cells were observed.
Statistical methods
SPSS13.0 statistical software was used for statistical analysis of all the test data. The non-parametric test, Wilcoxon test, was used for comparison between the groups. P<0.05 was considered statistically significant, and P<0.01 was highly statistically significant.
Results
MRI results
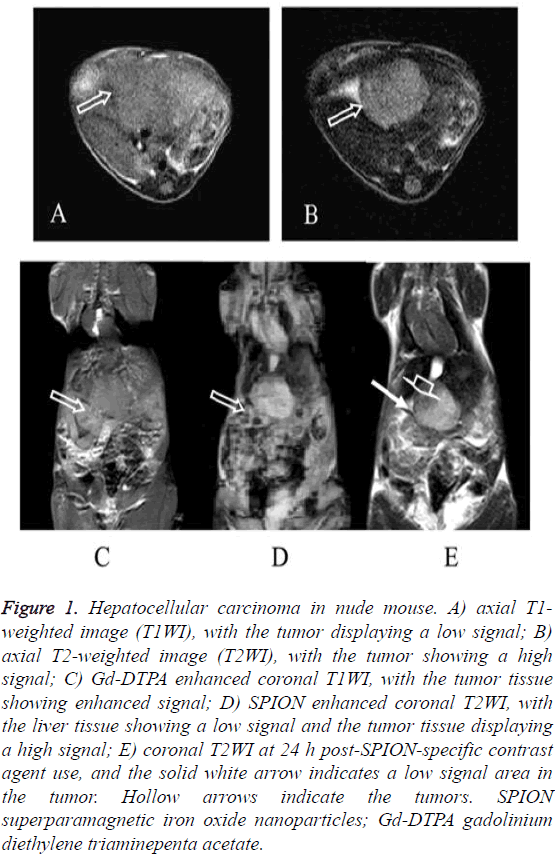
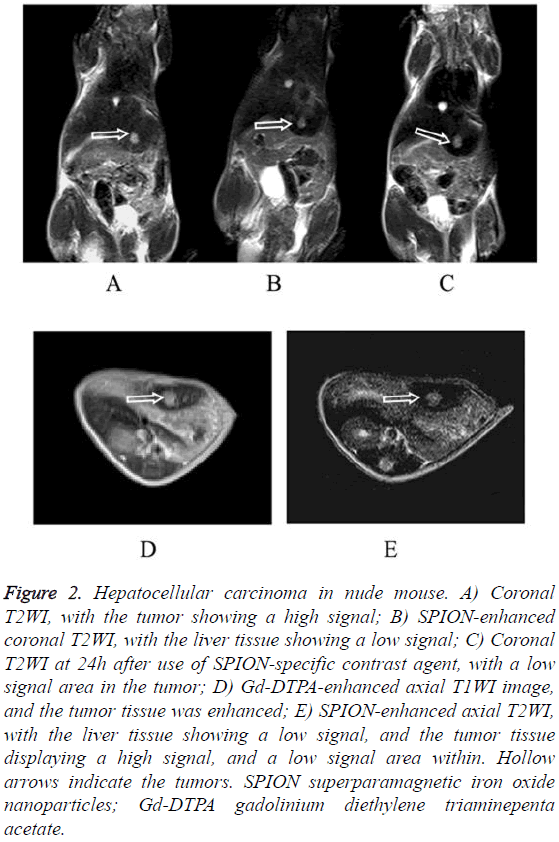
The HepG2 and the SMMC7721 groups included a total of 40 samples subjected to 1.5 T MRI at 1 w, 2 w, 3 w, 4 w, 5 w and 6 w after tumor transplantation. Two attending physicians analyzed and recorded the test results, respectively. The tumor showed slightly lower signal on unenhanced T1WI and slightly higher signal on T2WI. The conventional Gd-DTPA contrastenhanced T1WI showed positive enhancement (Figure 1), SPION-enhanced T2WI and T2* sequences manifested a significantly lower signal in liver tissue and a relative higher signal in tumor tissue (Figure 2). Liver tissue 24 h after targeting human primary liver cancer survivin gene with siRNA combined with SPIO still displayed low-signal changes. Compared with examination immediately after SPION enhancement, low-signal negative enhancement was found on T2WI, specifically (Figures 1-3).
Figure 1: Hepatocellular carcinoma in nude mouse. A) axial T1- weighted image (T1WI), with the tumor displaying a low signal; B) axial T2-weighted image (T2WI), with the tumor showing a high signal; C) Gd-DTPA enhanced coronal T1WI, with the tumor tissue showing enhanced signal; D) SPION enhanced coronal T2WI, with the liver tissue showing a low signal and the tumor tissue displaying a high signal; E) coronal T2WI at 24 h post-SPION-specific contrast agent use, and the solid white arrow indicates a low signal area in the tumor. Hollow arrows indicate the tumors. SPION superparamagnetic iron oxide nanoparticles; Gd-DTPA gadolinium diethylene triaminepenta acetate.
Figure 2: Hepatocellular carcinoma in nude mouse. A) Coronal T2WI, with the tumor showing a high signal; B) SPION-enhanced coronal T2WI, with the liver tissue showing a low signal; C) Coronal T2WI at 24h after use of SPION-specific contrast agent, with a low signal area in the tumor; D) Gd-DTPA-enhanced axial T1WI image, and the tumor tissue was enhanced; E) SPION-enhanced axial T2WI, with the liver tissue showing a low signal, and the tumor tissue displaying a high signal, and a low signal area within. Hollow arrows indicate the tumors. SPION superparamagnetic iron oxide nanoparticles; Gd-DTPA gadolinium diethylene triaminepenta acetate.
The sensitivity of MRI was higher than 90% following enhanced examination of liver tumors with diameter ≥ 0.2 cm. MRI positive rate was the highest at 24 h post-enhancement with human HCC survivin gene siRNA combined SPION. The high probability of positive rate was almost consistent with the pathology. In liver tumors with diameter <0.2 cm, the MRI scanner operated at a low field strength (1.5 T), without the surface coil. Therefore, the image resolution was affected. However, the tumor detection rate 24 h post-enhancement with HCC survivin gene siRNA combined SPION was still a magnitude higher compared to the level immediately after SPIO enhancement or conventional Gd-DTPA contrastenhanced examination. At a tumor diameter <0.1 cm, the scanning techniques were affected by the spatial resolution of MR, and failed to delineate the tumors. However, tumor cells were visible under a microscope.
SPSS13.0 statistical software was used for non-parametric statistical analysis of all the test data. The results are as follows: Analysis of the total tumor detection rate indicated that the differences between unenhanced MRI and immediately after SPION enhancement, conventional Gd-DTPA contrastenhanced examination and examination 24 h after enhancement with HCC survivin gene siRNA combined with SPION was highly statistically significant (P<0.01). The difference between conventional Gd-DTPA contrast-enhanced examination and examination 24 h after enhancement with HCC survivin gene siRNA combined SPION was statistically significant (P<0.05). The difference between immediate examination after SPIO enhancement and conventional Gd- DTPA contrast-enhanced examination and examination 24 h post-HCC survivin gene siRNA combined SPION was not statistically significant (P>0.05) (Tables 1-5).
| Unenhanced | SPION enhanced | SPION24 enhanced* | GD enhanced ** | Pathology | |
|---|---|---|---|---|---|
| Positive | 9 | 23 | 25 | 22 | 35 |
| Probably positive | 7 | 5 | 4 | 3 | 0 |
| Negative | 24 | 12 | 11 | 15 | 5 |
| Sensibility | 0.257 | 0.657 | 0.714 | 0.629 | 1 |
Table 1: Total tumor detection rate of MRI post- implantation.
| Unenhanced | SPION enhanced | SPION24 enhanced* | GD enhanced ** | Pathology | |
|---|---|---|---|---|---|
| MRI detection rate of tumors with diameter ≥ 0.2 cm | |||||
| Positive | 9 | 20 | 21 | 20 | 22 |
| Probably positive | 6 | 2 | 1 | 2 | 0 |
| Negative | 7 | 0 | 0 | 0 | 0 |
| Sensibility | 0.409 | 0.909 | 0.955 | 0.909 | 1 |
| MRI detection rate of tumors with diameter <0.2 cm | |||||
| Positive | 0 | 2 | 4 | 2 | 13 |
| Probably positive | 1 | 3 | 3 | 1 | 0 |
| Negative | 12 | 8 | 6 | 10 | 0 |
| Sensibility | 0 | 0.154 | 0.308 | 0.154 | 1 |
**Conventional Gd-DTPA contrast agent enhanced. SPION superparamagnetic iron oxide nanoparticles; Gd-DTPA gadolinium diethylene triaminepenta acetate; siRNA small interfering RNA; GD gadolinium diethylene.
Table 2: MRI detection rate of tumors.
| SPION-Unenhanced | SPION24-unenhanced | GD-Unenhanced | SPION24 - SPION | GD-SPION | GD-SPION24 | |
|---|---|---|---|---|---|---|
| Z | -3.729 | -4.041 | -3.314 | -1.342 | -1.414 | -2.111 |
| Asymp. Sig. (2-tailed) | 0.000 | 0.000 | 0.001 | 0.180 | 0.157 | 0.035 |
Table 3: Statistical analysis of total tumor detection rate.
| SPION-Unenhanced | SPION24-unenhanced | GD-Unenhanced | SPION24-SPION | GD-SPION | GD-SPION24 | |
|---|---|---|---|---|---|---|
| Z | -3.145 | -3.272 | -3.020 | -1.414 | 0.000 | -1.414 |
| Asymp. Sig. (2-tailed) | 0.002 | 0.001 | 0.003 | 0.157 | 1.000 | 0.157 |
Table 4: Analysis of tumor detection (diameter ≥ 0.2 cm).
| SPION- Unenhanced |
SPION24- unenhanced |
GD- Unenhanced |
SPION24- SPION | GD- SPION | GD- SPION24 | |
|---|---|---|---|---|---|---|
| Z | -2.070 | -2.460 | -1.414 | -0.577 | -1.633 | -1.667 |
| Asymp. Sig. (2-tailed) | 0.038 | 0.014 | 0.157 | 0.564 | 0.102 | 0.096 |
Table 5: Detection of tumor with diameter <0.2 cm.
In case of tumors with diameter ≥ 0.2 cm, the difference in detection rates between unenhanced MRI and immediately after SPION enhancement, conventional Gd-DTPA contrastenhanced examination, and examination at 24 h post-HCC survivin gene siRNA combined with SPIO was highly statistically significant (P<0.01). The difference between conventional Gd-DTPA contrast-and immediately after SPIO enhancement and examination 24 h post-enhancement with HCC survivin gene siRNA combined with SPIO was not statistically significant (P>0.05). The difference in detection rate between immediate examination after SPIO enhancement and examination 24 h after HCC survivin gene siRNA combined with SPIO was not statistically significant (P>0.05).
In case of tumors with diameter <0.2 cm, the difference in detection rate between unenhanced MRI and immediately after SPIO enhancement and post-SPIO enhancement and 24 h after HCC survivin gene siRNA combined with SPIO was remarkably statistically significant (P<0.05). The difference in detection rate between unenhanced MRI and conventional Gd- DTPA contrast-was not statistically significant (P>0.05). The difference between conventional Gd-DTPA contrast- and immediately after SPIO enhancement and 24 h after enhancement with HCC survivin gene siRNA combined with SPIO was not statistically significant (P>0.05). The difference in detection rate immediately after SPIO enhancement and 24 h after HCC survivin gene siRNA combined with SPIO was not statistically significant (P>0.05).
Discussion
In this nano-MRI study of T2WI negative contrast-enhanced molecular imaging of the tumors, siRNA specifically entered tumor cells to produce RNA interference, and superparamagnetic iron oxide particles were transported into the tumor cells with a high degree of target gene expression. T2WI in tumor tissue 24 h after tail vein injection combined with siRNA specific contrast agent, showed punctate lowsignal area, confirming successful MR molecular imaging of survivin genes in human primary liver cancer. All the tumors in this study were micro-hepatocellular carcinoma with a maximum diameter <1 cm. The difference in tumor detection rate 24 h after siRNA-specific contrast-enhancement and conventional Gd-DTPA contrast-enhanced examination was statistically significant (P<0.05). For tumors larger than 0.2 cm, although the difference in tumor detection rate 24 h after siRNA-specific contrast enhancement and conventional Gd- DTPA contrast-enhanced examination was statistically significant, it was not so (P>0.05) 24 h after siRNA-specific contrast enhancement. The typical low signal found on T2WI in the tumor tissues was still persistent.
Due to the limited spatial resolution of MRI, in this study, the detection rate for tumors <0.2 cm was still not high. The difference between siRNA-specific contrast and conventional Gd-DTPA contrast media was not statistically significant (P>0.05). The results had some limitations mainly associated with the limited sample size. In addition, the intravenously injected chemosynthetic siRNA was easily decomposed by RNA enzymes, unlike retroviruses, which continuously produce siRNA, indicating that RNA interference in vivo caused no significant tumor growth or changes. MRI reporter genes do not need a tissue sample. Representative MRI reporter gene model systems include melanin system, transferrin system, metal binding chimera or fusion peptide systems, and using 31P MRS creatine as a reporter gene for hepatic expression, as well as a recently designed special MR contrast agent showing gene expression by β-galactosidase enzymolysis [4,5]. However, in these systems, specific gene transfection was first conducted in vitro followed by imaging using special MR contrast agents. To image the abnormally expressed genes in tumor cells was not accomplished, and therefore, not amenable to clinical application.
In this study, the abnormally high expression of genes in human primary liver cancer cells was investigated to identify the characteristic MRI imaging profile. The approach enables early qualitative diagnosis of HCC. By improving the siRNA transfection mechanisms combined with transferrin receptor antibodies, it is possible to increase the transport efficiency of superparamagnetic iron oxide nanoparticles (SPION) into tumor cells [6,7]. MR molecular imaging probes, therefore, play a key role in cancer detection and diagnosis [8]. Compared with other transfection agents, PLL was costeffective [9]. Combining siRNA with PLL non-specifically increases cellular endocytosis. Resovist has a particle size of about 50~80 nm [10]. Resovist combines with PLL and siRNA to facilitate entry into cells by endocytosis. SiRNA targeted the increased expression of survivin genes in tumor cells through RNA interference. By effectively transporting Resovist complexed with PLL and siRNA into tumor cells, we have demonstrated the feasibility of using nano-MRI as a promising strategy for the early detection of metastatic hepatocellular carcinoma in situ. The methodology needs further clinical testing and regulatory approval.
References
- Tuschl T, Borkhardt A. Small interfering RNAs: a revolutionary tool for the analysis of gene function and gene therapy. Mol Interv 2002; 2: 158-167.
- Li KA, Zhang F, Ma YJ. Preliminary study on SPIO molecular probe labeled nude mice lung adenocarcinoma transplantation tumor MRI and its pathology. Chinese J Med Imaging Technol 2005; 21: 1655-1658.
- Liao W, Chen LY, Guo QY. Clinical study on liver-specific MRI contrast agent-feridex. J Clin Radiol 2001; 20: 251-255.
- Gong Y. Molecular imaging contrast agent research progress (review). Int J Med Radiol 2006; 29: 294-298.
- Blasberg R. Imaging gene expression and endogenous molecular processes: molecular imaging. J Cereb Blood Flow Metab 2002; 22: 1157-1164.
- Bulte JW, Kraitchman DL. Iron oxide MR contrast agents for molecular and cellular imaging. NMR Biomed 2004; 17: 484-499.
- Schäfer R, Kehlbach R, Wiskirchen J. Transferrin receptor upregulation: in vitro labeling of rat mesenchymal stem cells with superparamagnetic iron oxide. Radiology 2007; 244: 514-523.
- Hussain T, Nguyen QT. Molecular imaging for cancer diagnosis and surgery. Adv Drug Deliv Rev 2014; 66: 90-100.
- Hwang H S, Hu J, Na K. Role of polymeric endosomolytic agents in gene transfection: a comparative study of poly(L-lysine) grafted with monomeric L-histidine analogue and poly(L-histidine). Biomacromolecules 2014; 15: 3577-3586.
- Wang S, Fang J, Zhang T. Magnetic resonance imaging targeting of intracranial glioma xenografts by Resovist-labeled endothelial progenitor cells. J Neuro-Oncol 2011; 105: 67-75.