ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 11
Molecular mechanism of microRNA-93 in suppressing the cell proliferation of cervical cancer by adjusting the EGFR/AKT signal pathway
Xue Dong* and Yan Wang
Department of Obstetrics and Gynecology, the Second Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan, PR China
- *Corresponding Author:
- Xue Dong
Department of Obstetrics and Gynecology
The Second Affiliated Hospital of Zhengzhou University, China
Accepted date: March 28, 2017
Purpose: This study aims to discuss and explore the influences of microRNA-93 on the cell proliferation of cervical cancer and its regulation mechanism.
Methods: Constructed microRNA-93 expression vectors were transfected into the HeLa cell line through lipofection, after which the expression of microRNA-93 in HeLa cells was detected through qRT-PCR. In addition, the influences of microRNA-93 on HeLa cell proliferation were detected through CCK-8. Western blot was used to analyse changes of the EGFR/AKT signal pathway in Hela cells after the overexpression of microRNA-93.
Results: Compared with the control group, microRNA-93 was effectively heterologously expressed in HeLa cells. The CCK-8 results demonstrated that heterologous overexpression of microRNA-93 obviously suppressed Hela cell proliferation. Western blot showed that heterologous overexpression of microRNA-03 inhibited the expression of the EGFR/AKT signal pathway.
Conclusions: The heterologous overexpression of microRNA-03 may inhibit the proliferation of cervical cancer cells. Hence, microRNA-93 can regulate the proliferation of cervical cancer cells by suppressing the EGFR/AKT signal pathway.
Keywords
Cervical cancer cell, MicroRNA-93, Cell proliferation, EGFR/AKT signal.
Introduction
Cervical cancer is one of the most common malignant tumors seen in females, ranking 2nd in female malignant tumors, and has severely threatened the life and health of female patients [1]. Early cervical cancer often shows no apparent symptoms, and is therefore difficult to diagnose; unfortunately, middle or late cervical cancer has a low curative ratio [2]. Hence, finding highly sensitive and specific diagnostic markers for early cervical cancer and its effective therapy plan is an urgent research task [3]. MicroRNAs are non-coding single-stranded small RNAs (~20 nt in length) that inhibit the expression of a target gene by binding to the target RNA 3’UTR. In addition, microRNAs participate in the genesis and progression of tumors and play an important role in tumor cell proliferation, apoptosis, and migration [4]. In tumor genesis, microRNAs play a role that is similar to that of an oncogene or an antioncogene. The present study aimed to discuss the molecular mechanism of microRNA-93 as it regulates the proliferation of cervical cancer cells, and further provide theoretical basis for the clinical treatment and prevention of cervical cancer.
Materials and Methods
Cell source
Human cervical HeLa cells were purchased from the Cell Resource Center, Shanghai Institute for Biological Sciences, China Academy of Science.
Reagents and instruments
The expression vector pEGFP-C1 was from Clontech, US; Taq DNA polymerase HindIII, restriction endonuclease BamHI, and AMV reverse transcriptase, among others, came from Takara Company; homologous recombination enzyme EZfusion was from Shanghai General Company; DH5α competence cells were from Beijing TransGen Biotech; lipofection reagent Lipofectamine® 3000 was from US Invitrogen; DMEM culture media, Fetal Bovine Serum (FBS), and trypsin came from Gibco; CCK-8 kit was from Shanghai Beyotime Biotechnology; clean bench and CO2 incubator came from Thermo Fisher; flow cytometer FACSCalibur came from the US BD Company; and the qRT-PCR instrument was from the ABI Company.
Construction of the expression vector
The location of microRNA-93 in the genome was specified through miRBase. The base sequence of mature microRNA-93 and its flank were obtained using the public database Ensembl. The corresponding primer was designed for the 150 bp fragment extended towards the upstream and downstream of mature microRNA-93 (Table 1). Taq DNA polymerase was used to obtain the corresponding PRC products. The purified target PCR bands were collected by gel extraction kit. The expression vector pEGFP-C was linearized through double digestion by HindIII and BamH. The purified PCR fragment was connected into the linearized vector pEGFP-C1 through EZfusion. After subsequent transformation and identification, the expression vector of microRNA-93 was constructed.
| Name | Sequence | |
|---|---|---|
| microRNA-93RT-PCR primer | Upstream: | AAGTGCTGTTCGTGCAGGT |
| Downstream: | CTCGGGAAGTGCTAGCTCA | |
| U6 RT-PCR primer | Upstream: | CGCGACAAGGCCAAGAT |
| Downstream: | GCTGCTCCACCTTCTTCTG | |
| microRNA-93 PC amplification primer | Upstream: | AGATCTCGAGCTCAAGCTAG |
| TCTCTGGCTGACTACATCACAG | ||
| Downstream: | TCTAGATCCGGTGGATCCC | |
| TACTC ACAAAACAGGAGTGGAATC | ||
Table 1: Nucleotide sequence.
Cell culture and lipofection
HeLa cells were cultured in DMEM media containing 10% FBS, and then placed in an incubator with 5% CO2 and 95% air at 37°C. MicroRNA-93 was overexpressed heterologously in HeLa cells by lipofection. By following the transfection procedures for Lipofectamine® 3000, pEGFP-C1 (Vector) and pEGFP-C1-microRNA-93 (miRNA) were transfected into HeLa cells, after which the cells were cultured in an incubator containing 5% CO2 and 95% air at 37°C for two days. Then, the expression of green fluorescent protein was observed using fluorescence microscope. Next, HeLa cells were collected, and RNA was extracted and reversed. Finally, the expression of microRNA-93 in HeLa cells was detected by qRT-PCR.
RNA extraction, reverse-transcription, and qRT-PCR detection
HeLa cells were collected centrifugally, and the RNAs were extracted by Trizol method. The extracted RNAs were quantified and then reversely transcripted to be cDNAs by AMV reverse-transcription enzyme. cDNAs were used as templates for qRT-PCR detection. The microRNA-93-specific primer for qRT-PCR detection was produced by Sangon Biotech (Table 1). The PCR reaction system consisted of the following: 2X SYBR mix; 5 and 0.2 μL for upstream and downstream primers (10 μM), respectively; and template cDNA 25 ng. The system was filled to 10 μL using sterilized water. The reaction conditions were 95°C for 1 min, and then denaturation at 95°C for 15 s, followed by annealing at 60°C for 15 s and extension at 72°C for 30 s for a total of 40 cycles. The fluorescence signals were collected once per cycle. Using the expression of U6 as reference, the relative expression of microRNA-93 was relatively quantified through the expression of 2-ΔΔt pairs of genes.
Detection of cell proliferation activity
HeLa cells transfected with vector and miRNA were separately cultured in 96-well plates, each containing 2000 cells. Each group of HeLa cells seeded in 10 wells and cultured at 37°C for 48 h. To five wells of each group, 10 μL of CCK-8 liquid was added for 2 h. Then, the ELISA instrument was used to detect absorbance at 450 nm. The relevant absorbance value between the miRNA group and the vector group was calculated by subtracting the detected value for the group with no added CCK-8 from that of the group with CCK-8.
Apoptosis detection by annexin V and PI markers
HeLa cells transfected with vector and miRNA were separately collected by centrifugal method, and then marked with Annexin V and PI, respectively, according to the procedures of the two kits. Then, flow cytometer was used to detect cells marked with Annexin V and PI for apoptosis analysis.
Cell cycle detection by PI staining
HeLa cells transfected with vectors and miRNA were separately collected by centrifugal method, fixed by 75% ethanol, and then stained by PI. The cell cycle was analysed using a flow cytometer.
Western blot
The Hela cells transfected with vector and miRNAs were separately collected by centrifugal method, to which RIPA lysis buffer was added. The mixture was placed on ice for 0.5 h, after which 2X loading buffer was added. After lysis, cells were treated with 10% SDS-PAGE gel electrophoresis and transformed to another membrane for 100 min at 200 mA constant current; these were then closed in 5% skimmed milk at room temperature for 1 h, after which EGFR, AKT or p- EGFR, and p-AKT primary antibody dilution were added. This was then incubated at 4°C for one night, washed by TBS thrice (5 min per wash), and the corresponding secondary antibody dilution was added. Next, it was incubated at room temperature for 1 h and then washed by TBS thrice (5 min per wash). Finally, HRP-chromogenic reagent was used to show the Western blot results. All the antibodies used in this test were purchased from American CST Company.
Results
Heterologous overexpression of microRNA-93 in HeLa cells
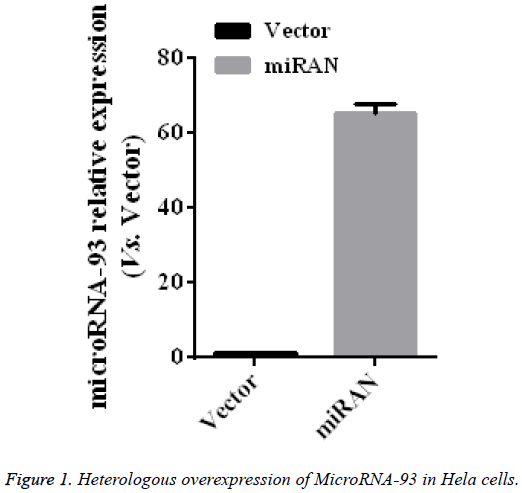
To identify the role of microRNA-93 in the cervical cancer HeLa cell line, EZfusion homologous recombination enzyme was used to construct the overexpression vector of microRNA-93. First, HeLa cells were transfected with pEGFPC1 (vector) and pEGFP-C1-microRNA-93 (miRNA) with lipofection method, cultured for 2 days, and then observed under a microscope. Most cells expressed GFT, indicating that the expression vectors were transfected into HeLa cells at high levels. After collecting the two groups, the microRNA-93 expressions in these two groups were measured by qRT-PRC, the results of which were compared with those of the vector group. The expression of microRNA-93 in the miRNA group obviously increased (p<0.01) by 65.5 ± 2.5 times that of the vector group (n=5, p<0.01) (Figure 1).
Influence of the heterologous overexpression of microRNA-93 on HeLa cell proliferation
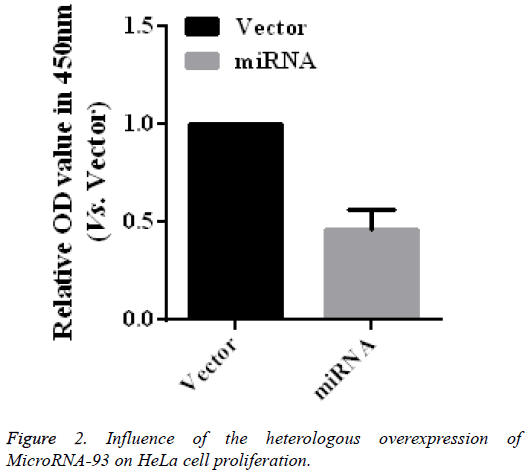
To further verify the role of microRNA-93 in modifying the proliferation of HeLa cells, HeLa cells transfected with vector and miRNA were cultured for 48 h, after which cell proliferation was detected by using CCK-8. As shown in the CCK-8 results (Figure 2), the relative absorbance of the microRNA-93 group at 450 nm was 0.46 ± 0.10 times that of the miRNA group (n=5, p<0.01), thereby indicating the heterologous overexpression of microRNA-93, which may significantly suppress HeLa cell proliferation.
Influence of the heterologous overexpression of microRNA-93 on Hela cell apoptosis
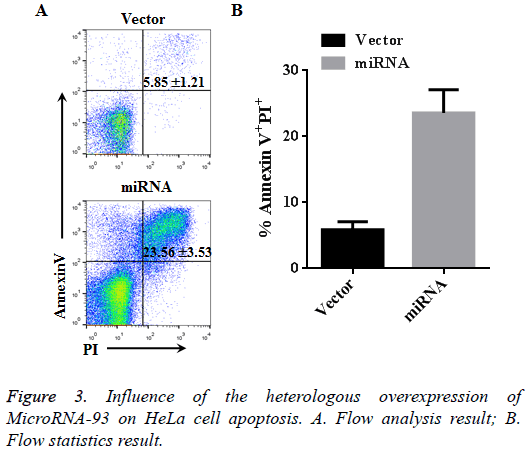
Hela cells transfected with vector and miRNA were cultured for 48 h, and apotosis was detected using Annexin V and PX markers. Based on the flow cytometer results, the cell apoptosis in the vector group (Annexin V+PI+) was 5.85% ± 1.21%, whereas that in the miRNA group with heterologously overexpressed microRNA-93 was 23.56% ± 3.53%. Therefore, the apoptosis of the miRNA group significantly increased compared with that of the vector group (n=3, p<0.01, Figure 3).
Influence of the heterologous overexpression of microRNA-93 on HeLa cell cycle
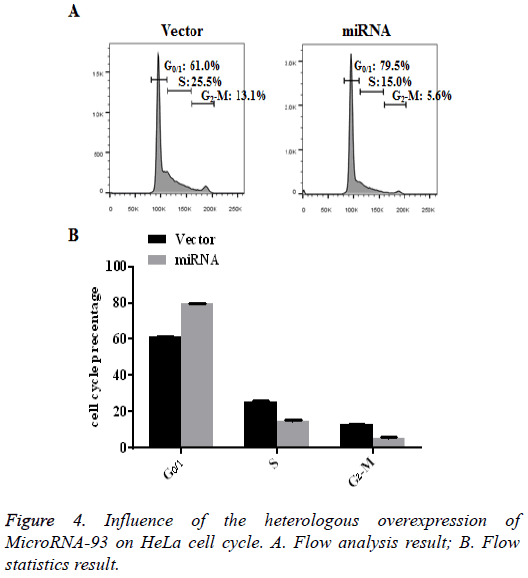
HeLa cells transfected with vector and miRNA were cultured for 48 h, after which the cell cycle of each group was detected by PI staining. The flow results showed that, compared with the vector group, most cells in the miRNA group with heterologously overexpressed microRNA-93 were arrested in G0/1 (79.5% ± 0.25%). The results are shown in Figure 4 (n=3, p<0.01).
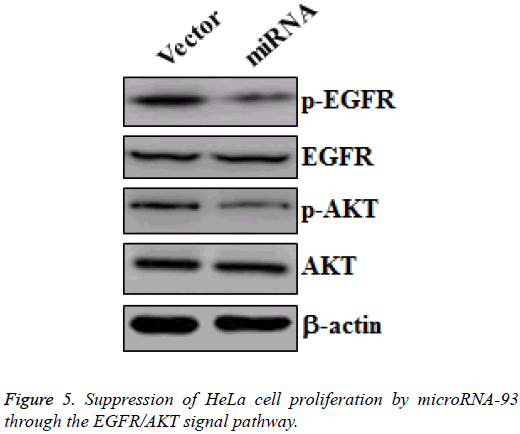
Suppression of HeLa cell proliferation by microRNA-93 through the EGFR/AKT signal pathway
MicroRNA-93 suppresses the proliferation of cervical cancer cell by promoting apoptosis and G0/1 cell cycle arrest. To reveal the mechanism that allows microRNA-93 to inhibit cervical cancer cell proliferation, we collected protein samples from HeLa cells transfected with vector and miRNA, and then cultured these for 48 h. Western blot was made to measure the changes of p-EGFR and EGFR, as well as the p-AKT and AKT in the two groups. HeLa cells transfected with miRNA demonstrated the significant downregulation of p-EGFR and p- AKT, whereas no obvious changes in EGFR and AKT were observed (Figure 5). Hence, microRNA-93 inhibited the proliferation of HeLa cells by suppressing the EGFR/AKT signal pathway.
Discussion
Cervical cancer is one of the most common female malignant tumors in the world. Patients with cervical cancer have a high mortality rate as this disease can severely threaten the health and life quality of females. Despite the growing maturity of surgery, chemotherapy, and radiotherapy treatments, no satisfactory treatment has been discovered for the treatment of late cervical cancer or metastasis. Fortunately, current advancements in molecular target treatment offer new ideas for the treatment of cervical cancer [5]. The current study’s findings prove that the effective heterologous overexpression of microRNA-93 in human cervical cells can significantly inhibit EGFR/AKT signal pathway expression, thus suppressing the proliferation of cervical cancer cells.
Previous studies have reported that the abnormal expression of microRNA is associated with the proliferation, apoptosis, invasion, and migration of cervical cancer cells [6]. MicroRNAs could be categorized into two types depending on specific roles, namely, carcinogenic microRNAs and anticarcinogenic microRNAs. Carcinogenic microRNAs, such as microRNA-361-5p and microRNA-21, are often highly expressed in cervical cancer cells to promote the progression of cervical cancer and participate in the development or invasion of cervical cancer cells, which are positively correlated with poor prognosis [7]. In comparison, anti-carcinogenic microRNAs, such as microRNA-138 and microRNA-195, can suppress the proliferation, migration, and invasion of cervical cancer cells. The current study has successfully constructed the overexpression vector for microRNA-93, which may be transfected into cervical cancer cells through lipofection and become highly expressed. The qRT-PCR results showed that microRNA-93 was effectively heterologously expressed in HeLa cells, indicating that microRNA-93 may be effectively expressed in human cells and can thus play further roles. The CCK-8 results directly proved that microRNA-93 obviously suppressed the proliferation of endometrial cancer cell. Moreover, by using flow cytometer to analyse apoptosis and cell cycle, the results showed that the overexpression of microRNA-93 can arrest cervical cancer cells at G0/1 cycle, thus promoting apoptosis. This study also revealed that microRNA-93 suppressed the proliferation of cervical cancer cells. This finding explains its downstream signal EGFR/AKT pathway. Furthermore, microRNA-93 plays a role similar to that of an anti-oncogene in cervical cancer cells. The anticancer function of microRNA-93 may have possible applications in gene therapy, which is one of the relatively popular directions for tumor treatment research. Gene therapy refers to the introduction of target genes at the DNA or RNA level of the target cell, and then inducing the genes to express and play specific bio-effects to help cure patients [8-10]. In this study, we have constructed the transient overexpression vector for microRNA-93. However, for further gene therapy options, the functions of the stable expression of microRNA-93 in cervical cancer should be explored.
Conclusion
This study revealed the microRNA mechanism involved in cervical cancer cell proliferation, thus contributing to possible microRNA-based cervical cancer treatments in the future. The study also offers theoretical basis for future research on individualized gene therapy for cervical cancer.
References
- Zhang JZ, Huang ZY, Wang JS, Wang WJ, Ren G, Qi JD. Bisphenol AF (BPAF) and 3, 3, 5, 5-Tetrabromobisphenol A (TBPA)-Induced colorectal cancer risk through inhibiting the activity of UDP-Glucuronosyltransferase (UGT) 1A8. Lat Am J Pharm 2017; 36: 416-420.
- Hu S, Tao R, Wang S, Wang C, Zhao X. MicroRNA-21 promotes cell proliferation in human hepatocellular carcinoma partly by targeting HEPN1. Tumour Biol 2015; 36: 5467-5472.
- Peralta-Zaragoza O, Deas J, Meneses-Acosta A, De la O-Gomez F, Fernandez-Tilapa G. Relevance of miR-21 in regulation of tumor suppressor gene PTEN in human cervical cancer cells. BMC Cancer 2016; 16: 215.
- Ghorab MM, Alsaid MS, Alqasoumi SI, Abdel-Kader MS. Semisynthesis of some novel urea, thiourea, carbamimidothioic acid and dihydrooxazole derivatives as a new class of anticancer agents. Lat Am J Pharm 2017; 36: 380-385.
- Li K, Zhou N. Lung cancer Drug-Drug Interaction (DDI) related with praeruptorin D. Lat Am J Pharm 2017; 36: 184-187.
- Elaiyaraja G, Kumaratharan N. An enhanced algorithm for removal of noise in CT scan image and 3D abdomen color video sequence through trimmed based filter. Biomed Res India 2017; 28: 512-518.
- Meng F, Henson R, Wehbejanek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007; 133: 647-658.
- Chinnasamy G, Subramani K, Srinivasan V. Green synthesis, characterization and anxiolytic, sedative and hypnotic activity of pyrimidine based diazepine derivatives. Biomed Res India 2017; 28: 525-531.
- Lin F, Ding R, Zheng S, Xing D, Hong W, Zhou Z, Shen J. Decrease expression of microRNA-744 promotes cell proliferation by targeting c-Myc in human hepatocellular carcinoma. Cancer Cell Int 2014; 14: 58.
- Chandrakar C, Sharma M. Qualitative features selection techniques by profiling statistical features of ECG for classification of heart beats. Biomed Res India 2017; 28: 571-576.