ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 12
Molecular identification of trypanosomes in tsetse flies trapped from Onicha Ugbo in Delta state of Nigeria
Onyekwelu KC1, Ejezie FE1, Eze AA1, Ikekpeazu JE1, Isaac C2, Ibegbu DM1 and Ogbunude POJ1*
1Department of Medical Biochemistry, Faculty of Basic Medical Sciences, University of Nigeria, Enugu-Campus, Nigeria
2Department of Zoology, Ambrose Alli University, Ekpoma, Nigeria
- *Corresponding Author:
- Ogbunude POJ
Department of Medical Biochemistry
Faculty of Basic Medical Sciences
University of Nigeria, Nigeria
Accepted date: April 28, 2017
The delta region of Southern Nigeria is believed to be endemic for animal trypanosomiasis. Occasionally, cases of suspected human trypanosomiasis based on clinical presentation have been reported, thus we undertook survey of tsetse flies from the area with the purposes of typing the trypanosomes in the locality and demonstrating the possibility that human trypanosomiasis may be present in the area if the vector Trypanosoma gambiense is seen. A total of 429 flies were trapped; all were of Glossina palpalis palpalis specie. Sixty or 14.0% of them were found to be infected by trypanosomes. The infection rates were 7.7%, 1.6%, 1.6% and 6.3% for Trypanosoma vivax, Trypanosoma brucei brucei, Trypanosoma simiae and Trypanosoma congolense respectively. Mixed infections in various combinations were observed. There was no evidence of human infective Trypanosoma gambiense. The presence of various species of animal trypanosomes even as mixed infections confirms that the area is indeed endemic for animal trypanosomiasis. The simultaneous occurrence of Glossina palpalis. palpalis and Trypanosoma brucei brucei, however, makes it imperative to monitor the area continuously for human trypanosomiasis.
Keywords
Trypanosomiasis, Trypanosomes, Tsetse flies, Onicha Ugbo, Delta state of Nigeria
Introduction
Trypanosomes are protozoa organisms of major public and economic importance in sub-Saharan Africa causing both human and animal trypanosomiasis [1,2]. The disease transmitted mainly by tsetse fly in this region seriously limits the use of large areas of land for livestock production [3]. The transmission cycle of the disease involves three elements: the mammalian reservoirs of the parasites, the tsetse fly that serves as cyclic vectors for transmission and the pathogenic parasite, the trypanosomes.
The human and animal trypanosomiasis is caused by many species of trypanosomes carried by over twenty species and subspecies of tsetse flies (Glossina spp.). The tsetse fly ingests blood meal from infected hosts and over a period of weeks (approximately 4 weeks) the ingested trypanosome undergoes morphological and physiological transformations in the alimentary tract and salivary glands to become infective metacyclic.
Tsetse fly is a key factor in trypanosomiasis epidemiology therefore identification of trypanosomes in the flies should be a good indicator of extent of problem in an area. The Delta State of Nigeria is believed to be a focus for animal trypanosomiasis and occasionally suspected human trypanosomiasis had been reported [4,5], hence detection and identification of pathogenic trypanosomes in tsetse flies in the area will highlight the risks to humans and animals.
The trypanosomes, T. congolense, T. vivax, T. simiae and T. b. brucei are together responsible for most of African animal trypanosomiasis or nagana of cattle [6]. Trypanosomiasis is a major constraint to livestock production in sub-Saharan Africa [7]. Animals kept in areas of moderate risk of trypanosomiasis have lower calving rates, lower milk yields, higher rates of calf mortality and require more frequent treatment with preventive and curative doses of trypanosomal drugs than animals kept in trypanosomiasis free zones [8]. Thus trypanosomiasis has a major impact on animal farming.
The purpose of this study was to detect and identify the pathogenic trypanosomes in tsetse fly circulating in Oniche Ugbo, a town in delta region of Delta State. The study will highlight the risk of trypanosomiasis in the study area and in addition will help in proper planning of control measures to allow animal farming in this area with vast arable land.
Methods
Study site
The study was conducted on tsetse flies collected from Onicha ugbo (5º 46’ N, 6º 35’ E), an area that is located in Delta State of South-South region of Nigeria (Figure 1). It has mixed vegetation ranging from mangrove thick forest to rain forest and grasslands. The inhabitants are mainly subsistence farmers rearing mainly pigs, sheep and goats.
Tsetse flies
The tsetse fly samples were collected in the geographical area described in study site using pyramidal traps. The traps were not baited. Flies were collected at intervals of twenty minutes to prevent possible escape of trapped flies. The tsetse flies were stored separately at room temperature in 1.8 ml tubes containing absolute alcohol until used for DNA extraction.
DNA extraction
The tsetse flies were individually squashed with micro mortar and pestle followed by DNA extraction with Bioneer AccuPrep Genomic DNA extraction kit according to the manufacturer’s instruction. Briefly, squashed fly was transferred into a 1.5 ml tube using a total of 200 μl sterile water, to which 200 μl of proteinase K (20 mg/ml) was added. The tube was incubated at 60°C for 10 min. After mixing the lysate with 100 μl of isopropanol and vortexing, the mixture was transferred into the upper reservoir of the binding column. The column was washed twice with wash buffers 1 and 2 and then centrifuged at 13,400 g for 1 min. The binding column was transferred to a new 2 ml tube and filtered using the wash buffers and then eluted for 5 min with 200 μl of elution buffer into the glass fiber of the binding column and centrifuged for another 1 min at 13,400 g. The eluate from this centrifugation contains the genomic DNA which was stored at 4°C for later analysis.
Species identification
Identification of different species of trypanosomes was performed by amplifying the internal transcribed spacer one (ITS1) of the ribosomal DNA using technique previously described [9-12]. This method detects eleven pathogenic Trypanosoma species in a single PCR with size differences of the products given as: Trypanozoon (T. b. brucei, T. evansi, T. b. gambiense and T. b. rhodesiense) 480 bp; T. congolense Savannah subgroup 700 bp; T. congolense Kilifi subgroup 620 bp; T. congolense Forest subgroup 710 bp; T. simiae 400 bp; T. simiae tsavo 370 bp; T. godfrey 300 bp and T. vivax 250 bp. Briefly, the amplification was performed in 20 μl reaction vessel containing 3.5 μl Master mix (Promega USA) and 0.5 μl each of primers ( ITS1 CF: 5’- CCGGAAGTTCACCGATATTG-3’; ITS1 BR: 5’- TTGCTGCGTTCTTCAACGAA-3’), 3.5 μl of sterile water and 3.0 μl of gDNA. The vessels containing the reactants were placed in the thermocycler and programmed as follows: an initial denaturation at 95°C for 300 s, was followed by 30 cycles of 95°C for 30 s, 58°C for 30 s and 72°C for 60 s with a final extension at 72°C for 300 s.
For the assay involving the differentiation of human infective Trypanozoon, primer specific for T. b. gambiense (TgsGP F: 5’-GCTGCTGTGTTCGGAAGAGC-3’, TgsGP R: 5’- GCCATCGTGCTTGCCTCTC-3’) was used [13-15]. The reaction mixture consisted of 2.5 μl 10X Super Taq buffer, 0.5 μl of primersi, 0.2 μl of 100 mM dNTP, 1 μl of Taq DNA polymerase, 0.75 μl of divalent Mg2+ cation, 14.55 μl of sterile distilled water and 5 μl of genomic DNA. Amplification condition was an initial denaturation at 95°C for 300 s followed by 37 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 30 s with final extension at 72°C for 300 s.
5 μl of the PCR products were electrophoressed in 1.5% agarose gel containing 0.1 μg of ethidium bromide per ml, and bands visualized using UV transillumination. The runs included 100 bp markers.
Results
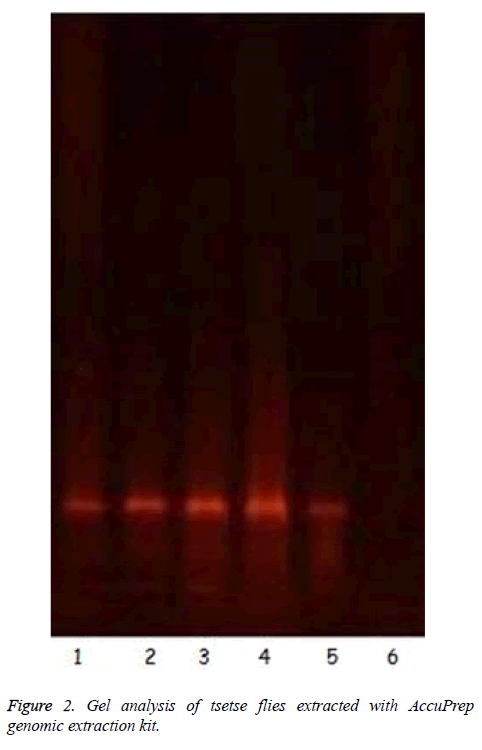
The results showed that all tsetse flies collected were G. p. palpalis. Isolation of gDNA from whole squashed fly with AccuPrep Genomic Extraction kit was interesting in the sense that the kit was designed for use in extraction of DNA from whole blood, leukocytes, mammalian tissues or cultured cells. It was successfully adapted for gDNA isolation from tsetse flies. The purity of the nucleic acids were checked by measuring the ratio of A260/A280 at neutral pH. All the values were in the range of 1.6 to 1.9. The incubation time of the squashed fly with proteinase K was critical; incubation less than or 5 min did not extract detectable DNA (Figure 2, lane 6). Ten minutes incubation was ideal to produce detectable DNA on 1.5% agarose gel as seen in Figure 2 (Lanes 1-5).
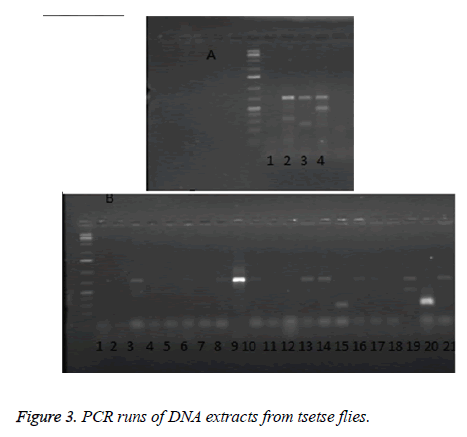
Figure 3 shows a representative result of PCR amplifications of gDNA isolates from tsetse fly using ITS1 BR and ITS1 CF primers. Mixed infections were evident in many of the flies, for example, in the upper panel (Figure 3A) there is evidence of mixed infections in Lanes 2 and 4. The lower panel, Figure 3B, shows that many of the flies were indeed infected. There is no observable pattern of mixing, any of the parasites coexisted, and however, T. vivax and T. congolense were seen more frequently either existing alone or in combination with each other or with other parasites.
Amplification of Trypanozoon with T. b. gambiense specific primers did not show positive result.
Discussion
Several species of tsetse flies are known to exist in nature of which about eleven species have been reported in Nigeria [16]. These species infest approximately 70% of the land mass of Nigeria, however, information on the biodiversity and distribution are scanty [17]. Generally, tsetse flies are grouped into three main subgroups depending on the environment they inhabit, thus the palpalis spp are mostly found in riverine areas, the morsitans spp. in the savannah vegetation and fuscipes spp. are forest-dwelling [18]. Our study showed that only G. palpalis was trapped in the study area. A similar study in northern Nigeria revealed that Wuya in north west of Niger State with savannah grassland was also colonized by G. palpalis while Yankari game reserve in Bauchi state, north East which also have savannah vegetation with patches of woodland hosts G. m. morsitans and G. tachnoides [19], thus there appears to be no exclusivity in tsetse flies distribution between riverine and savannah vegetation of Nigeria.
The main issue we addressed in our study was identification of trypanosomes in the flies, hence, our decision to use DNA probes based on repetitive DNA elements. This technique allowed the identification of subgenus of trypanosomes [20-22]. The major trypanosome species associated with dissemination of animal trypanosomes, namely T. vivax, T. congolense, T. simiae and T. brucei were all identified in our study area. The most frequently encountered trypanosomes were mostly those of the cattle, T. vivax, T. congolense, and T. brucei. The trypanosomes were found either singly or mixed in the tsetse flies. Mixed infections have been previously documented in multiple trypanosome species [23,24] and were interpreted to mean either due to high probability of chronic infection in susceptible host or high chances of trypanosome infections by tsetse flies or even both. Theoretically, it may be concluded that the presence of these trypanosomes in the tsetse flies implied active transmission of the organisms, however, the presence of parasite DNA does not necessarily translate to the presence of mature, transmissible organisms and therefore is not a direct indicator of risk [25]. We, thus, can only conclude that the molecular identification allowed us to identify trypanosomes species circulating in the study area.
The samples positive for Trypanozoon were subjected to further PCR analysis as discussed in Materials and Methods using primers specific for T. b. gambiense. None of the samples were positive for human infective trypanosomes. Despite the absence of human infective trypanosomes, it is still important to continue active monitoring of the area since livestock and wild animals are known to act as reservoirs for human African trypanosomes [26-28].
Conclusion
The presence of various species of animal trypanosomes even as mixed infections suggests that the area is endemic for animal trypanosomiasis. There is no evidence of human trypanosomes, however, the simultaneous occurrence of G. p. palpalis and T. b. brucei makes it imperative to continue monitoring for human trypanosomiasis in the locality. The high rates of T. congolense and T. vivax suggest a high potential risk of animal trypanosomiasis in livestock’s in the area.
Competing Interests
The authors declare that there is no conflict of interest regarding the publication of this paper.
References
- Truc P, Bailey JW, Doua F, Laveissiere C, Godfrey DG. A comparison of parasitological methods for diagnosis of Gambian trypanosomiassis in an area of low endemicity in Cote dIvoire. Trans Soc Roy Soc Trop Med and Hyg 1998, 88: 215-218.
- Barrett MP, Burchmore RJ, Stich A, Lazzari JO, Frasch AC. The trypanosomiases. Lancet 2003; 362: 1469-1480.
- Van den Bossche P. Some general aspects of the distribution and epidemiology of bovine trypanosomosis in southern Africa. Int J Parasitol 2001; 31: 592-598.
- Enwezor FNC, Ukah JCA. Advanced trypanosomiasis (sleeping sickness) in a child: case report. Nig J Parasitol 2000, 21: 143-146.
- Airauhi L, Unuigbe E, Airauchi OD. Human sleeping sickness (SS) in Nigeria: knowledge, attitude and beliefs in a focus in the Abraka belt, Delta of Nigeria. Afri J Clin Exp Microbiol 2001; 2: 6-9.
- Food, agriculture and food security. The global dimension 2002, WFS02/Tech/Advanced Unedited Version. Rome FAO 2002; 19-28.
- Vanhamme L, Pays E. The trypanosome lytic factor of human serum and the molecular basis of sleeping sickness. Int J Parasitol 2004; 34: 887-898.
- Swallow B. 2000 impacts of trypanosomiasis on African agriculture. PAAT Tech Scientific FAO Rome 1999; 1-46.
- Nakayima J, Nakao R, Alhassan A, Mahama C, Afakye K, Sugimoto C: Molecular epidemiological studies on animal trypanosomes in Ghana. Parasit Vect 2012, 5: 217-223.
- Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM. The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res 2005; 95: 186-192.
- Desquesnes M, McLaughlin G, Zoungrana A, Davila AMR. Detection and identification of Trypanosoma of African livestock through a single PCR based on internal transcribed spacer 1 of rDNA. Int J Parasitol 2001; 31: 610-614.
- Cox A, Tilley A, McOdimba F, Fyfe J, Eisler M. A PCR based assay for detection and differentiation of African trypanosome species in blood. Exp Parasitol 2005; 111: 24-29.
- Masiga DK, Smyth AJ, Hayes P, Bromidge TJ, Gibson WC. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol 1992; 22: 909-918.
- Gibson W. Molecular characterization of field isolates of human pathogenic trypanosomes. Trop Med Int Health 2001, 6: 401-406.
- Radwanska M, Claes F, Magez S, Magnus E, Péres-Morga D, Pays E, Buscher P. Novel primer sequences for a polymerase chain reaction-based detection of Trypanosoma brucei gambiense. Am J Trop Med Hyg 2002; 67: 289-295.
- Baldry DAT, Riodan K. A review of 50 years entomology of insect-borne diseases of veterinary importance in Nigeria, with special reference to tsetse flies and trypanosomiasis. Fifty years applied entomology in Nigeria. Conference of the Entomological Society of Nigeria, University of Ife 1967; 43-55.
- Dede PM, Halid I, Ommogun GA, Uzoigwe NR, Njoku CI, Daniel AD. Current tsetse and trpanosomiasis situation on Jos Plateau, Nigeria. Epizootiological factors that may enhance transmission and spread. Rev Elev Med Vet Pay 2005; 58: 31-35.
- Wamwiri FN, Changasi RE. Tsetse flies (Glossina) as vectors of human african trypanosomiasis: a review. Biomed Res Int 2016; 2016: 6201350.
- Issac C, Ciosi M, Hamilton A, Scullion KM, Dede P, Igbinosa IB, Nmorsi OPG, Masinga D, Turner CMR. Molecular identification of different trypanosome species and subspecies in tsetse flies of northern Nigeria. Parasit Vec 2016; 9: 301-307.
- Gibson WC, Dukes P, Gashumba JK. Species-specific DNA probes for the identification of African trypanosomes in tsetse flies. Parasitology 1988; 97: 63-73.
- Moser DR, Cook GA, Ochs DE, Baily CP, McKane MR, Dolson JE. Detection of Trypanosoma congolense and T. brucei subspecies of DNA amplification using the polymerase chain reaction. Parasitol 1989; 99: 57-66.
- Masiga DK, Smyth AJ, Hayes P, Bromidge TJ, Gibson WC. Sensitive detection of trypanosomes in tsetse flies by DNA amplification. Int J Parasitol 1992; 22: 909-918.
- Mekata H, Konnal S, Simuunza M, Chembensofu M, Kano R, Witola WH, Tembo M, Chitambo H, Inoue N, Onuma M, Ohashi K. Prevalence and source of trypanosome infections in field-captured vector flies (Glossina Paiiidipes) in Southern Zambia. J Vet Med Sci 2008; 70: 923-928.
- Malele II, Magwisha HB, Nyingilili HS, Mamiro KA, Rukambile EJ. Multiple Trypanosoma infections are common amongst Glossina species in the new farming areas of Rufiji district, Tanzania. Parasit Vectors 2011; 4: 217.
- Auty HK, Picozzi K, Malele I, Torr SJ, Cleaveland S, Welburn S. Using molecular data for epidemiological inference: assessing the prevalence of Trypanosoma brucei rhodesiense in tsetse in Serengeti, Tanzania. PLOS Negl Trop Dis 2012; 6: 1501.
- Simo G, Asonganyi T, Nkinin SW, Njiokou F, Herder S. High prevalence of Trypanosoma brucei gambiense group 1 in pigs from the Fontem sleeping sickness focus in Cameroon. Vet Parasitol 2006; 139: 57-66.
- Njiokou F, Laveissiere C, Simo G, Nkinin S, Gre`baut P, Cunny G, Herder S. Wild fauna as a probable animal reservoir for Trpanosoma brucei gambiense in Cameroon. Infect Genet Evol 2006; 6: 147-153.
- Njioku F, Nimpaye H, Simo G, Njitchoang GR, Asonganyi T, Cuny G, Herder S. Domestic animals as potential reservoir hosts of Trypanosoma brucei gambiense in sleeping sickness foci in Cameroon. Parasite 2010; 17: 61-66.