ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 5
Molecular docking studies of kirenol a traditional Chinese medicinal compound against rheumatoid arthritis cytokine drug targets (TNF-?, IL-1 and IL-6)
Department of Rheumatology and Clinical Immunology, ZhuJiang Hospital of Southern Medical University, Guangzhou 510282, PR China
#These authors contributed equally to this work
- *Corresponding Author:
- Qing-hong Yu
Department of Rheumatology and Clinical Immunology
ZhuJiang hospital of Southern Medical University, PR China
Accepted on March 03, 2016
Rheumatoid Arthritis (RA) an autoimmune multifactorial disease impairing the quality of life of the diseased is a challenging prospect in terms of treatment. The disease has been treated from ancient times and from their wisdom, we have taken one traditional Chinese medicinal compound named kirenol. The compound is from herbal extract of Herba Siegesbeckiae and in this study we are exploring its inhibiting potential of cytokine target i.e. TNF-α, IL-1 and IL-6 using Auto Dock tool. In our study we have used Auto Dock 4.2 an Insilico tool to check the effect of the kirenol on three important cytokine target i.e. TNF-α, IL-1 and IL-6. The molecular docking approach using AutoDock 4.2 tool with default parameters was used to calculate the binding energies. All the three cytokines showed spontaneous binding with kirenol, varying from a ΔG range of -6.75 to -2.68 Kcal/mole. The results generated showed IL-6 to be the best target for kirenol. Both, its binding energy and number of interactions is higher when compared to that of other two.
Keywords
Rheumatoid arthritis, TNF-α, IL-1, IL-6, Kirenol, Molecular docking.
Introduction
Rheumatoid Arthritis (RA) is an autoimmune disease resulting in chronic inflammation disorder that affects many tissues and organs in almost 1% of the adult population [1]. RA is characterized by the chronic inflammation of synovial membrane in affected joints, ultimately leading to dysfunction of joint [2]. The impaired quality of life, joint deformity, morbidity and mortality are the features of RA [3,4]. The etiology of RA is unknown, however progress have been made in development of new therapeutics [5].
Since 1998 new biological drugs for RA have been introduced and approved for treatment [6], these drugs are known as Disease Modifying Anti-Rheumatic Drugs (DMARDs), which have role in curbing the underlying process of the disease and they have become the mainstay of the treatment of RA [7]. In our institute (ZhuJiang hospital) for the treatment of RA a new compound called kirenol is being used to a great effectiveness. Kirenol, a Traditional Chinese Medicine (TCM) is an extract of Herba Siegesbeckiae botanical name Siegesbeckia pubescens Mak a traditionally used herb found abundantly in China and practiced for the treatment of arthritis, hypertension, malaria, neurasthenia and snakebite [8].
As the compound possess anti-inflammatory properties and is a known TCM against RA, so we used it for molecular docking analysis against the cytokine target involved in RA. The main three known targets of RA disease are TNF-α, IL-1 and IL-6 [9-14] were considered for the study.
Material and Methods
Protein preparation
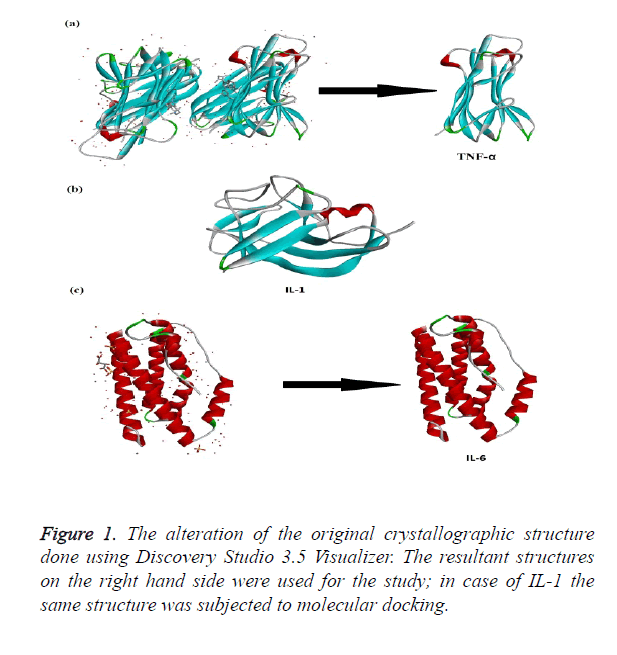
The crystallographic structures of proteins TNF-α, IL-1 and IL-6 were retrieved from protein database website (www.pdb.org). TNF-α crystallographic structure bearing PDB ID: 2AZ5 was selected for the study [15] the partial crystallographic structure of TNF-α covered the protein structure from amino acid 10-157. IL1 crystallographic structure bearing PDB ID: 2ILA from position 117-271 [16]. The third structure of Il-6 (PDB ID: 1ALU) covered from 28 - 212 of the protein structure [17]. All the three structures were trimmed according to the needs of the study using Discovery Studio 3.5 Visualizer [18]. The energy minimization of the trimmed protein was cried out by SPDB Viewer.
Ligand structure
For kirenol compound, the coordinates were taken from NCBI PubChem compound database (www.pubchem.ncbi.nlm.nih.gov/) bearing Chem ID: 15736732. The structure was in molfile format and was changed to PDB format using Discovery Studio 3.5 Visualizer.
Molecular docking
The docking of kirenol into TNF-α, IL-1 and IL-6 crystallographic structure was performed using AUTODOCK 4.2 [19]. The AUTODOCKTOOLS [20] were used for preparing the protein for docking, the polar hydrogens, partial charges and Gastegier charges were added using these tools. The flexible torsion of the ligand was assigned same for all three proteins; the AutoGrid tool was used to implement the auto grid file, different for each protein. The grid parameters set for each run are in Table 1. The AutoDock was finally used for blind docking of kirenol into the crystallographic structure of TNF-α, IL-1 and IL-6.
| Target Protein | Grid Size A˚ (x , y , z) | Coordinates (x, y, z) |
|---|---|---|
| TNF-α | 118, 106, 94 | -25.6, 67.1, 42.6 |
| IL-1 | 126, 90, 90 | 0.8, -0.6, -1.1 |
| IL-6 | 122, 110, 100 | 2.6, -19.9, 8.8 |
Table 1. The Grid parameters fallowed for blind docking of kirenol into TNF-α, IL-1 and IL-6.
Visualization
The docking log file of each run was used to generate top three binding poses of each protein, based on the binding energy of kirenol. The saved poses were visualized using Discovery Studio 3.5 Visualizer.
Results
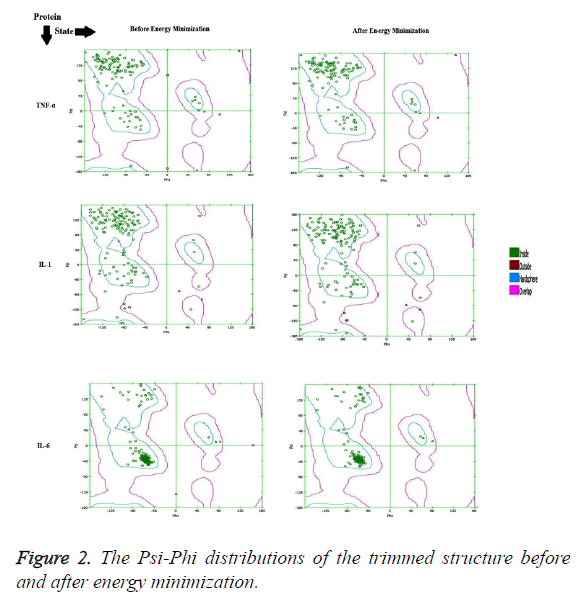
The crystallographic structure of TNF-α constituted: four subunits, water and the ligand. The structure was trimmed to a single subunit as shown in Figure 1a. The IL-1 crystallographic structure was used as such for the blind docking (Figure 1b). The IL-6 structure was trimmed and the modifications are shown in Figure 1c. The Psi-Phi distribution of the trimmed protein structures before and after energy minimizations are depicted in Figure 2, showing the structures after energy minimization within the ramachandran limit.
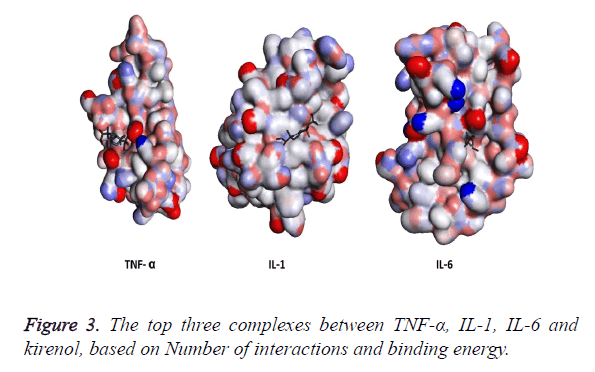
The three structures after modification were then subjected to AutoDock and the top three results generated by kirenol are shown in Table 2. On the basis of binding energy the top interaction of kirenol and TNF-α protein is the most spontaneous having the binding energy of -6.75 Kcal/mol. The top three poses of each protein were saved in pdbqt format and were then analyzed for the interaction between the protein and TCM using Discovery Studio 3.5 Visualizer. On the basis of the number of interactions between the target protein and Kirenol, three complexes were selected (Figure 3), which are described in Table 3.
| Target Protein | Rank | ΔG (Kcal/mole) | No. of Interactions |
|---|---|---|---|
| TNF-α | 1 | -6.75 | One |
| 2 | -4.56 | One | |
| 3 | -4.17 | Two | |
| IL-1 | 1 | -4.9 | - |
| 2 | -3.89 | Three | |
| 3 | -2.68 | - | |
| IL-6 | 1 | -6.72 | Two |
| 2 | -6.53 | One | |
| 3 | -4.52 | One |
Table 2. Top three binding energies of each TNF-α, IL-1 and IL-6 with kirenol, the results are generated by AutoDock4.2.
| Target Protein | ΔG (Kcal/mole) | Interactions |
|---|---|---|
| TNF-α | -4.17 | Kirenol: O23-TNF-Al: ILE136: O (2.6 A˚) |
| TNF-Al:LEU26:N - Kirenol:O24(2.7 A˚) | ||
| IL-1 | -3.89 | Kirenol: O24 - IL-1:LEU92: O (2.8 A˚) |
| IL-1:GLN38:HE22 - Kirenol:O24(2.1 A˚) | ||
| IL-1:GLN38:HE22 - Kirenol:O23(2.3 A˚) | ||
| IL-6 | -6.72 | Kirenol: O19 - IL-6:ASN63: O (2.1 A˚) |
| IL-6:LEU64:N - Kirenol:O19(2.8 A˚) |
Table 3. Top three complexes with their binding energies and interaction pattern, the results are generated by Discovery Studio 3.5 Visualizer.
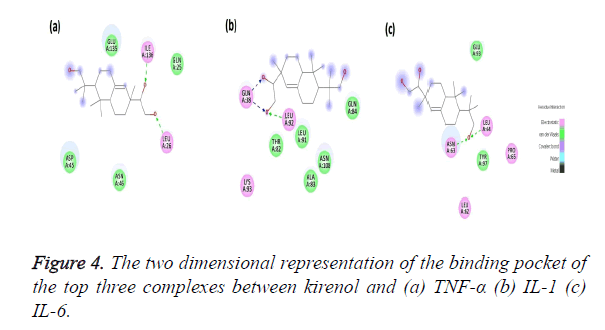
The Kirenol-TNF-α protein interaction (covalent or non-covalent) and the number of hydrogen bonds formed have been displayed in Figure 4a. The best complex has a ΔG of -4.75 Kcal/mol and forms two hydrogen bond with ILE136 and LEU26 of TNF-α binding pocket. The binding pocket in this case is of following amino acids GLN25, LEU26, ASP45, ASN46, GLU135 and ILE136. The O23 position of the kirenol is interacting with ILE136 and the bond formed between them has a distance of 2.6 A˚. The other interaction is between the O24 position of the kirenol with LEU26 and the distance here is 2.7 A˚.
The second most spontaneous interaction of kirenol-IL1 protein is the most stable, with the number of hydrogen bonds being three. Its binding pocket comprises of GL38, THR82, ALA83, GLN84, LEU91, LEU92, LYS93 and ASN108. It has ΔG of -3.89 Kcal/mol and out of three hydrogen bonds GLN38 forms two with O23 and O24 of kirenol with a distance of 2.3 and 2.1A˚ respectively. The other hydrogen bond is between O24 atom of kirenol with a distance of 2.8A˚. Figure 4b shows the mapping of this kirenol-IL1 complex.
Figure 4c shows the last interaction, most spontaneous of the top three selected with a ΔG of -6.72 Kcal/mol. The binding pocket of this complex comprises of following amino acids viz. LEU62, ASN63, LEU64 PRO65, GLU93 and TYR 97. Out of them kirenol forms two hydrogen bonds with ASN63 and LEU64, the O atom at the 18th position of kirenol is forming the hydrogen bonds with ASN63 and LEU64 with a distance if 2.1 and 2.8 A˚.
Discussion
RA is a disease with unknown pathophysiology leading to joint destruction and ultimately disability. The disease can be targeted via. various important biological pathways involved, the important drug able target molecules of these pathways can be targeted and in our study we are looking into the TCM compound kirenol, a traditionally used RA drug and are exploring it cytokine based targets TNF-α, IL-1 and IL-6 by In silico approaches. Natural origin compounds have tremendous potential in blocking immunomodulators [21], our study also looks into the prospective activity of resveratrol on RA by targeting immunomodulators. Kirenol has been long reported to have an effect on RA [22], however the mechanistic mechanism of action was yet to be elucidated. In summary, the results of this study provide a comprehensive assessment of immunosuppressive pathways by which Kirenol can induce a potent protective effect against RA.
Acknowledgement
The authors would like to take an opportunity to thank the management of ZhuJiang hospital for providing the funding and computational support to carry forward this work.
References
- Lawrence RC, Helmick CG, Arnett FC, Deyo RA, Felson DT, Giannini EH, Heyse SP. Estimates of the prevalence of arthritis and selected musculoskeletal disorders in the United States. Arthritis Rheum 1998; 41: 778-799.
- Pincus T, Callahan LF. Reassessment of twelve traditional paradigms concerning the diagnosis, prevalence, morbidity and mortality of rheumatoid arthritis. Scand J Rheumatol 1989; 79: 67-96.
- Pincus T, Callahan LF. What is the natural history of rheumatoid arthritis? Rheum Dis Clin North Am 1993; 19: 123-51.
- Puolakka K. Work Capacity and Productivity Costs in Early Rheumatoid Arthritis: A Five-Year Prospective Study (dissertation). (Cited 2013, March 12). Helsinki: Academic Dissertation University of Helsinki, 2005.
- McInnes IB, Schett G. The Pathogenesis of Rheumatoid Arthritis. Mechanisms of Disease. N Engl J Med 2011; 365: 2205-2219.
- Fernández-Cruz E, Alecsandru D, Rodríguez-Sainz C. Introduction to biological drugs. Actas Dermosifiliogr 2008; 99: 2-6.
- Saag KG, Teng GG, Patkar NM, Anuntiyo J, Finney C, Curtis JR, Paulus HE, Mudano A. American College of Rheumatology 2008 recommendations for the use of non-biologic and biologic disease-modifying antirheumatic drugs in rheumatoid arthritis. Arthritis Rheum 2008; 59: 762-784.
- Pharmacopoeia of the People's Republic of China 2005. People's Medical Publishing House, 2005.
- Okamoto H, Hoshi D, Kiire A, Yamanaka H, Kamatani N. Molecular targets of rheumatoid arthritis. Inflamm. Allergy Drug Targets 2008; 7: 53-66.
- Williams RO, Feldmann M, Maini RN. Anti-tumor necrosis factor ameliorates joint disease in murine collagen-induced arthritis. Proc Natl Acad Sci USA 1992; 89: 9784-9788.
- Klareskog L, van der Heijde D, de Jager JP, Gough A, Kalden J, Malaise M. Therapeutic effect of the combination of etanercept and methotrexate compared with each treatment alone in patients with rheumatoid arthritis: double-blind randomised controlled trial. Lancet 2004; 363: 675-681.
- Keystone EC, Kavanaugh AF, Sharp JT, Tannenbaum H, Hua Y, Teoh LS, Fischkoff SA, Chartash EK. Radiographic, clinical, and functional outcomes of treatment with adalimumab (a human anti-tumor necrosis factor monoclonal antibody) in patients with active rheumatoid arthritis receiving concomitant methotrexate therapy: A randomized, placebo-controlled, 52-week trial. Arthritis Rheum 2004; 50: 1400-1411.
- Dinarello CA. Interleukin-1, interleukin-1 receptors and interleukin-1 receptor antagonist. Int Rev Immunol 1998; 16: 457-499.
- Madhok R, Crilly A, Watson J, Capell HA. Serum interleukin 6 levels in rheumatoid arthritis: correlations with clinical and laboratory indices of disease activity. Ann Rheum Dis 1993; 52: 232-234.
- He MM, Smith AS, Oslob JD, Flanagan WM, Braisted AC, Whitty A, Cancilla MT. Small-molecule inhibition of TNF-alpha. Science 2005; 310: 1022-1025.
- Graves BJ, Hatada MH, Hendrickson WA, Miller JK, Madison VS, Satow Y. Structure of interleukin 1 alpha at 2.7-A resolution. Biochemistry 1990; 29: 2679-2684.
- Somers W, Stahl M, Seehra JS. 1.9 A crystal structure of interleukin 6: implications for a novel mode of receptor dimerization and signaling. EMBO J 1997; 16: 989-997.
- Accelrys Software Inc., Discovery Studio Modeling Environment, Release 3.5, San Diego: Accelrys Software Inc 2012.
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated Docking Using a Lamarckian Genetic Algorithm and Empirical Binding Free Energy Function. J Comput Chem 1998; 19: 1639-1662.
- Sanner MF, Duncan BS, Carrillo CJ, Olson AJ. Integrating computation and visualization for biomolecular analysis: an example using python and AVS. Pac Symp Biocomput 1999: 401-412.
- Zhou W, Cai JF, Yuan F, Ma M, Yin F. In silico targeting of interleukin-6 by natural compounds. Bangladesh J Pharmacol 2014; 9: 371-376.
- Wang ZM, Zhu SG, Wu ZW, Lu Y, Fu HZ, Qian RQ. Kirenol upregulates nuclear annexin-1 which interacts with NF-κB to attenuate synovial inflammation of collagen-induced arthritis in rats. J Ethnopharmacol 2011; 137: 774-782.