ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 4
Isolation and identification of lactic acid bacteria (Lactobacillus plantarum YS2) from yak yogurt and its probiotic properties
Yu Qian1,2,3,4#, Xingyao Long5#, Yanni Pan1,2, Guijie Li1,2,3,4* and Xin Zhao1,2,3,4*
1Chongqing Collaborative Innovation Center for Functional Food, Chongqing University of Education, Chongqing, P.R. China
2Chongqing Engineering Research Center of Functional Food, Chongqing University of Education, Chongqing, P.R. China
3Chongqing Engineering Laboratory for Research and Development of Functional Food, Chongqing University of Education, Chongqing, P.R. China
4College of Biological and Chemical Engineering, Chongqing University of Education, Chongqing, P.R. China
5College of Food Science, Southwest University, Chongqing 400715, P.R. China
#These authors have contributed equally to this work
- *Corresponding Authors:
- Xin Zhao
Chongqing Collaborative Innovation Center for Functional Food
Chongqing University of Education
PR China - Guijie Li
Chongqing Collaborative Innovation Center for Functional Food
Chongqing University of Education
PR China
Accepted date: November 25, 2017
DOI: 10.4066/biomedicalresearch.29-17-3418
Visit for more related articles at Biomedical ResearchIn this study, a lactic acid bacterium was isolated and identified from Yushu in China, and its probiotic properties were determined. After cultured in MRS medium, a bacterium was identified as lactic acid bacteria by staining and microscopic observation. And the lactic acid bacteria was further identified as Lactobacillus plantarum by physiological and biochemical tests. The lactic acid bacterium was also identified as Lactobacillus plantarum by 16sDNA determination, and the lactic acid bacteria was named Lactobacillus plantarum YS2 (LP-YS2). The acid resistant activity of LP-YS2 (68.05%) were higher than that of Lactobacillus bulgaricus (LB, 30.35%). LP-YS2 also showed the higher bile salt resistant activities (19.85%, 15.01% and 8.35%) than those of LB (3.08%, 1.87% and 1.44%) in 0.3%, 0.5% and 1.0% bile salt. The hydrophobicity results also showed that LP-YS2 (72.35%) had the higher hydrophobicity than LB (45.67%). LP-YS2 had the good values in food and bio pharmaceutical development.
Keywords
Lactic acid bacteria, Yak yogurt, Probiotic properties, 6S rDNA, DNA
Introduction
Yaks are mainly distributed in the Qinghai Tibet Plateau and its adjacent alpine areas. There are 3 million 900 thousand Yaks in Tibet, accounting for 30% of the total number of Yaks in the world [1]. Yak provides livestock, meat, fur and other livestock products for Tibetan herdsmen, which is an important economic source of local people [2]. Compared with normal milk, the yak milk is rich in protein, essential amino acids, fat, lactose and minerals [3]. With fresh yak milk as raw material, the traditional fermented yak yogurt is kind of flavored dairy product of a low alcohol content, which is made from various lactic acid bacteria and yeasts and so on, as a complex microbial system with many excellent characteristics of lactic acid bacteria. The research found that there are Lactobacillus delbrueckii, Lactococcus, Enterococcus, Bacillus and other lactic acid bacteria in solidified yak yoghurt in pastoral area of Yushu Tibetan Autonomous Prefecture, Qinghai, China [4]. Twelve kinds of lactic acid bacteria are found by analysis of Tibet traditional fermented cheese through PCR-DGGE method (Naizha) in lactic acid bacteria, including L. paracasei, fermentation of Lactobacillus, Lactobacillus plantarum, among which Lactobacillus delbrueckii is the dominant bacteria [5].
As general term for beneficial microorganisms, probiotics are good for the parasitifer as microbial colonization in the human intestinal tract and reproductive system, which can produce the exact health effects so as to improve micro ecological balance of parasitifer, and play a useful role in the activity [6]. Beneficial bacteria or fungi in the human and animals mainly include Clostridium butyricum, Lactobacillus, Bifidobacterium, Actinomycetes and yeast etc. [7]. Probiotics have been now widely used in bioengineering, industrial and agricultural, food safety and life and health fields. With the rapid development of fermented dairy products, the development of probiotic products with better physiological functions has become an important direction for the development of dairy industry [8]. Lactic acid bacteria with gastric and intestinal juice and intestinal absorption capacity can be used as potential probiotics for further development [9]. Li et al. [10] found one strain of Lactobacillus helveticus MB2-1 was isolated from traditional fermented milk. The study showed that the strain had good antioxidant activity in vitro, the research on probiotic lactic acid bacteria of milk curd in Inner Mongolia king series showed that Lactobacillus plantarum M1-2 and Lactobacillus rhamnosus M6-1 have good probiotic properties with potential application value through the simulation of artificial gastric juice, bile salt and hydrophobic test [11].
In this study, the natural fermented yak yogurt from Yushu area of Gansu province, China were isolated, and the traditional combination of physiological and biochemical experiments and 16S rDNA gene sequence analysis is conducted to identify one strain of lactic acid bacteria, and its probiotic potential in vitro is studied at the same time, which provide the basic information for the development and utilization of resources of lactic acid bacteria in yak yogurt.
Materials and Methods
Isolation and purification of lactic acid bacteria
The natural fermented yak yoghurt was adopted from herdsman family in Qinghai Tibet Plateau in Yushu, Qinghai, China. The solution we prepared contains 1 mL natural fermented yak yoghurt which was diluted to 10-7 in a sterile saline for a 10 fold gradient. The 10-5, 10-6 and 10-7 natural fermented yak yoghurt diluent was added on culture dish of 15 mL MRS medium (containing 5% CaCO3) it then cultured in the constant temperature incubator (Biochemical incubator BI-150A, STIK, Shanghai, China) for 72 h. After colony formation, the colonies of bacteria which had calcium dissolving ring were inoculated on skim milk medium for 48 h (30°C), then streak inoculated on MRS agar medium for 48 h (30°C), above two steps were repeated thrice until the pure colonies were obtained.
Identification of lactic acid bacteria
The pure strain was streak cultured on MRS medium for 48 h (30°C), the colony size and shape were observed and tested by many different ways such as by using a microscope (BX43, Olympus, Tokyo, Japan), the gram staining, catalase test, exercise test, hydrogen sulfide test, gelatin liquefaction test, nitrate reduction test, indole test, litmus milk test, methyl red test, V.P. test, 6.5% NaCl growth test, growth temperature test (10°C, 15°C, 45°C, 60°C for 30 min), pH gradient test and sugar alcohol fermentation test were all measured with high precautions.
DNA of lactic acid bacteria extraction
Liquid culture of lactic acid bacteria (2 mL) was centrifugated at 12000 r/min for 1 min, the supernatant fluid was discarded and the microbial biomass was collected. The DNA of lactic acid bacteria was extracted using the DNA extraction kit (Tiangen Biotech (Beijing) Co., Ltd., Beijing, China).
DNA of lactic acid bacteria determination
The extracted DNA solution was subjected to agarose gel electrophoresis (gel concentration 0.8%, voltage 100V, electrophoresis 60 min), the gel was observed using gel imaging system (6200, Tanon, Shanghai, China). The concentration of DNA extraction was determined by using a micro ultraviolet spectrophotometer (Nano300, Allsheng, Hangzhou, Zhejiang, China). The PCR gene was amplified by 27F (F), AGAGTTTGATCCTGGCTCAG and 1492R (R) GGCTACCTTGTTACGACTT, and using the 16SrDNA sequence of Lactobacillus it was amplified and sequenced (SimpliAmp Thermal Cycler, Thermo Fisher Scientific, Inc., Waltham, MA, USA).
The experimental microorganism strains
Lactobacillus bulgaricus (LB, CCTCC AB 200048) was purchased from CCTCC.
Acid tolerance activity
The lactic acid bacteria bacterial culture (5 mL) was collected by centrifugation (1,000 × g, 10 min). The collected bacterial pellet was re-suspended using a sterile saline (5 mL), and following which 1 ml suspension was mixed with 9 mL sterile artificial gastric juice (0.2% NaCl, 0.35% pepsin, pH 3.0) and incubated in a thermostatic oscillator at 37°C and 300 rpm. Subsequently, sample (200 µL) was pipetted well and incubated with a Lactobacillus MRS Agar plate at 37°C for 3 h. After 0 and 3 h later, the number of colony forming units (CFUs) was counted and calculated the bacterial survival rate, the survival in pH 3.0 artificial gastric juice was calculated according to formula: viable count at 3 h (CFU/mL) / viable count at 0 h (CFU/mL) × 100% [12].
Bile salt tolerance activity
According to the inoculation amount of 2%, the lactic acid bacteria solution was inoculated into MRS-THIO medium MRSTHIO medium (MRS medium was added with 0.2% sodium acetate) containing 0.0%, 0.3%, 0.5% and 1.0% cattle bile salt, respectively. After incubation at 37°C for 24 h, the blank medium was used as control, all the media were determined at 600 nm, and the growth in bile salt was calculated according to formula: bile salt containing culture medium OD600 nm/blank culture medium OD600 nm × 100% [12].
Hydrophobicity
The surface hydrophobicity of the strain was determined by BATH (carbon hydrocarbon adhesion method). The mycelia were collected at 5 mL and centrifuged at 3000 r/min for 10 min. 5 mL PBS (50 mmol/L, pH 6.5) of buffer mycelium was washed for 2 times with. PBS buffer was used as blank control, and the concentration was adjusted by PBS buffer, to make the OD560 value about 1.4 mL of bacterial fluid with concentration adjusted was put into 10 mL of sterile centrifugal tube, with 0.8 mL of xylene, while there is no xylene in the control group. The oscillation is 30 s, the pause is 10 s, and the oscillation is 30 s later. Then, standing for 5-10 min for layer, take the lower aqueous phase, with PBS buffer fluid as the blank control to test OD560 the value of nm. The hydrophobic rate (A0 and A are nm values of OD560 of bacteria fluid before and after mixing with xylene) are calculated according to the formula: (A0-A)/A0 × 100% [12].
Results
Lactobacillus plantarum colonial morphology
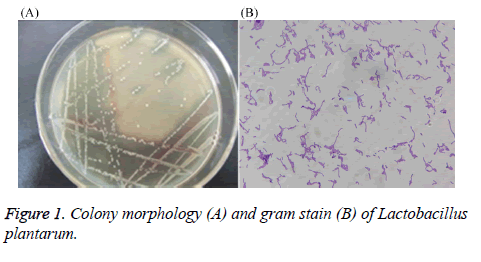
After performing MRS culture medium, the strain showed neat white, round, edge, centrally slightly raised, a diameter of about 1.6 mm, the surface is thick and opaque, smooth (Figure 1). Through gram staining, the stain also showed gram positive, long rod-shaped, arranged in clusters or arranged in chains, with darker staining. Therefore, preliminary identification confirmed that the stain might be Lactobacillus.
Physiological and biochemical test results of Lactobacillus
The physiological and biochemical test results of Lactobacillus were showed in Tables 1-3, the stain was identified as Lactobacillus plantarum.
| Text | |
|---|---|
| 10°C, 30 min growth | + |
| 15°C, 30 min growth | + |
| 45°C, 30 min growth | + |
| 60°C, 30 min growth | - |
| pH 4.5 growth | + |
| pH 9.6 growth | +w |
| Anaerobic growth | +w |
| Motility | - |
| Symbols: +, positive strain; negative strain; +w, weakly positive strain. | |
Table 1: Physiological test result of Lactobacillus.
| Text | ||
|---|---|---|
| Contact enzyme test | - | |
| Oxidase test | - | |
| Hydrogen sulfide test | - | |
| Gelatin liquefaction test | - | |
| Nitrate reduction test | - | |
| Indole test | - | |
| Glucose gas production test | - | |
| Benzidine test | - | |
| Litmus milk test | Acid production | + |
| Decolorization | + | |
| Freezing | + | |
| Peptonization | - | |
| Arginine producing ammonia test | - | |
| Casein decomposition test | - | |
| Symbols: +, positive strain; -, negative strain. | ||
Table 2: Biochemical test result of Lactobacillus.
| Sugar alcohol | |
|---|---|
| Amygdalin | + |
| Arabinose | + |
| Cellobiose | + |
| Aesculin | + |
| Fructose | + |
| Galactose | + |
| Glucose | + |
| Gluconate | + |
| Lactose | + |
| Maltose | + |
| Mannite | + |
| Seminose | + |
| Melizitose | - |
| Melibiose | + |
| Raffinose | + |
| Rhamnose | - |
| D-ribose | + |
| Salicin | + |
| Sorbierite | + |
| Sucrose | + |
| Fucose | + |
| Xylose | - |
| Symbols: +, positive strain; -, negative strain. | |
Table 3: Sugar alcohol fermentation test result of Lactobacillus.
DNA determination of Lactobacillus
The PCR amplification of 16sDNA gene results showed that negative control had no banding, the amplified had no pollution (Figure 2). The lane was a length of about 1500 bp the specificity of the amplified fragment, expected amplified fragment length (Figure 3). After sequence determination and matching gene sequence in Genbank, the sequencing result was GTGCAATGGCGGCTTGCCTAATACATGCAAGTCGAACGAACTCTGGATTGATTGGTGCTTGCATCATGATTTACATTTGAGGGAGTGGCGAACTGGTGAGTAACACGTGGGAAACCTGCCCAGAAGCGGGGAATAACACCTGGAAACAGATGCTATACCGCATAACAACTTGGACCGCATGGTCCGAGnTTGAAAGATGGCTTCGGCTATCACTTTTGGATGGTCCCGCGGCGTATTAGCTAGATGGTGGGGTAACGGCTCACCATGGCAATGATACGTAGCCGACCTGAGAGGGTAATCGGCCACATTGGGACTGACTCACGGCCAAACTCCTACGGGAGGCAGCAGTAGGGAATCTTCCACAATGGACGAAGTCTGATGGAGCAACGCCGCGTGAGTGAAAAGGGTTTCGGCTCGTAAACTCTGTTGTTAAAGAAGAACATATCTGAGAGTAACTGTTCAGGTATTGACGGTATTTAACAGAAAGCCACGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGTGGCAAGCGTTGTCCGGATTTATTGGGCGTAAAGCGAGCGCAGGCGGTTTTTTAAGTCTGATGTGAAAGCCTTCGGCTCAACCGAAGAAGTGCATCGGAAATGGGAAACTTGAGTGCAGAAGAGGACAGTGGAACTCCATTGTAGCGGTGAAATGCGTAGATATATGGAAGAACACCAGTGGCGAAGGCGCTGTCTGGTCTGTAACTGACGCTGAGGCTCGAAAGTATGGTAGCAAACAGGATTAGATACCCTGGTATCCATACCGTAAACGATGAATGCTAAGTGTTGGAGGGTTTCCGCCCTTCAGTGCTGCAGCTAACGCATTAAGCATTCCGCCTGGGGAGTACGGCCGCAAGGCTGAAACTCAAAGGAATTACGGGGGCCCGCACAAGCGGTGGAGCATGTGGTTTAATTCGAAGCTACGCGAAGAACCTTACCAGGTCTTGACATACTATGCAAATCTAAGAGATTAGACGTTCCCTTCGGGGACATGGATACAGGTGGTGCATGGTTGTCGTCAGCTCGTGTCGTGAGATGTTGGGTTAGTCCCGCACGAGCGCAACCCTTATTATCAGTTGCCAGCATTAAGTTGGGCACTCTGGTGAGACTGCCGGTGACAAACGGAGGAAGGTGGGGATGACGTCAAATCATCATGCCCCTTATGACCTGGGCTACACACGTGCTACAATGGATGGTACAACAGTTGCGAACTCGCGAGAGTAAGCTAATCTCTTAAAGCCATTCTCAGTTCGGATTGTAGGCTGCAACTCGCCTACATGAAGTCGGAATCGCTAGTAATCCGGATCAGCATGCCGCGGTGAATACGTTCCCGGGCCTTGACACACCGCCCGTCACACCATGAGAGTTTGTAACACCCAAAGTCGGTGGGGTAACCTTTTAGGAACCAGCCGCCTAAGGGGGACAGATGATTAGGGTGGGT (Lactobacillus plantarum strain CIP 103151). The isolation and identification of lactic acid bacteria was named Lactobacillus plantarum YS2 (LP-YS2), and this stain was preserved in China Center for Type Culture Collection (CCTCC, Wuhan, China) also we can introduce the preservation number which was M2013512.
Acid resistant activity of Lactobacillus plantarum YS2 and Lactobacillus bulgaricus
The colony counts of LP-YS2 at 0 and 3 h were more than LB, and following exposed to the artificial gastric juice, LP-YS2 exhibited higher gastrointestinal survival ability than that of LB (Table 4).
| Strain | 0 h colony count (CFU/mL) | 3 h colony count (CFU/mL) | Acid resistant activity (%) |
|---|---|---|---|
| LP-YS2 | 2.66 × 108 | 1.81 × 108 | 68.05 ± 7.39 |
| LB | 2.57 × 108 | 0.78 × 108 | 30.35 ± 5.74 |
| LP-YS2: Lactobacillus plantarum YS2; LB: Lactobacillus bulgaricus. | |||
Table 4: Acid resistant activity of Lactobacillus plantarum YS2 and Lactobacillus bulgaricus.
Bile salt resistant activity of Lactobacillus plantarum YS2 and Lactobacillus bulgaricus
In the 0.3%, 0.5% and 1.0% bile salt, LP-YS2 showed the greater bile salt resistant activity than that of LB (Table 5).
| Strain | Growth in bile salt (%) | ||
|---|---|---|---|
| 0.3% | 0.5% | 1.0% | |
| LP-YS2 | 19.85 ± 2.26 | 15.01 ± 1.77 | 8.35 ± 0.52 |
| LB | 3.08 ± 0.25 | 1.87 ± 0.22 | 1.44 ± 0.20 |
| LP-YS2: Lactobacillus plantarum YS2; LB: Lactobacillus bulgaricus. | |||
Table 5: Bile salt resistant activity of Lactobacillus plantarum YS2 and Lactobacillus bulgaricus.
Hydrophobicity of Lactobacillus plantarum YS2 and Lactobacillus bulgaricus
As shown in Table 6, the hydrophobicity of LP-YS2 was 72.35%, it was higher that of LB (45.67%). The hydrophobicity of LP-YS2 was 1.58 times than LB.
| Strain | Hydrophobicity (%) |
|---|---|
| LP-YS2 | 72.35 ± 4.21 |
| LB | 45.67 ± 3.86 |
| LP-YS2: Lactobacillus plantarum YS2; LB: Lactobacillus bulgaricus. | |
Table 6: Hydrophobicity of Lactobacillus plantarum YS2 and Lactobacillus bulgaricus.
Discussion
The identification methods of lactic acid bacteria can be divided into phenotype identification and genotype identification. The advantages of 16S rDNA sequence analysis and denaturing gradient gel electrophoresis (DGGE) technology in the classification and identification of lactic acid bacteria are becoming more and more obvious [4]. Lactic acid bacteria have been identified from traditional dairy products in Tibet, China, by using the methods of homology analysis of genus specific PCR, 16S rDNA sequence, and species specific PCR and NEB cutter analysis [13]. Other study has been done to identify lactic acid bacteria in Mongolian and Kazakh yogurt in Xinjiang, China by using traditional physiological and biochemical experiments and 16S rDNA gene sequence analysis [14].
Probiotics pass through the stomach before entering the gut, so the ability to fight gastric juice is the primary condition for probiotic screening [15]. Usually, the pH of gastric juice in human body is about 3, and the digestion time is between 1-3 h. The viability of each strain was evaluated by lactic acid bacteria digestion in the pH 3 artificial gastric juice for 3 h, and the lactic acid bacteria with good tolerance were screened out [12]. In this study, the tolerance of 4 strains of Lactobacillus in simulated gastric juice was studied. The survival rates of M5AL and J23AL were high; G15AL and T3AL lost their activity after exposure to 3 h in gastric juice [16]. Researchers have conducted experiments on the tolerance of Lactobacillus Isolated from Inner Mongolia koumiss samples in simulated gastric capacity, in addition to the L. casei Zhang with high survival rate, other experimental strains were less than 60% [17]. Therefore, the lactic acid bacteria isolated from yak yogurt were good in acid resistance. The difference in acid resistance of different lactic acid bacteria is related to proton pump mechanism, acid tolerance reaction mechanism, macromolecular protection and repair, alkali generation, etc. In addition, regulators, cell density and biofilm may also affect the acid resistance of lactic acid bacteria [18].
When probiotics enter the intestinal tract, the bile salts in the small intestine inhibit the probiotics. Therefore, the tolerance of the strains to bile salts is often one of the important indicators for screening probiotics [19]. Generally, the concentration of bile salts in human body fluctuates within the range of 0.03% to 0.3%. Studies have shown that the survival rate of lactic acid bacteria more than 70% in the artificial gastric juice can grow in different concentrations of bile salt, and the growth efficiency decreases with the increase of bile salt concentration. The reason is that high concentration bile salt can make the membrane permeability change, membrane protein dissociation, eventually lead to intracellular material flow, some cell death [20]. In the screening of probiotics in vitro, the surface hydrophobicity of bacteria reflects the ability of bacteria colonization in vivo, as with strong hydrophobic ability, lactic acid bacteria have good adhesion and strong colonization ability [21].
Conclusions
For lactic acid bacteria as the main probiotics, its probiotic properties have been an important basis for scholars to study probiotics. Studies have shown that probiotic lactic acid bacteria can regulate intestinal flora balance, inhibit cholesterol absorption, antioxidant, anti-tumor and cancer prevention and so on. In this study, the isolation and identification of a strain of lactic acid bacteria through physiological and biochemical tests and 16S rDNA gene sequence analysis further conduct experiment on the anti-traditional, artificial gastric juice, bile salt resistance and hydrophobicity, which showed the lactic acid bacteria of LP-YS4 having good anti artificial gastric juice, bile salt resistant ability and strong hydrophobicity. Therefore, this lactic acid bacteria has good probiotic potential with good value in food and bio pharmaceutical development.
Acknowledgement
The present research was supported by Chongqing Research Program of Basic Research and Frontier Technology (cstc2016jcyjA0339), Chongqing Engineering Research Center for Functional Food (cstc2015yfpt_gcjsyjzx0027), Research Project of Chongqing University of Education (KY2015TBZC), and the Program for Innovation and Team Building at the Chongqing Institute of Higher Education (CXTDX201601040), China.
References
- Zhong JC, Chen ZH, Zhao SJ, Xiao YP. Classification of ecological types of the Chinese yak. Acta Ecol Sinica 2006; 26: 2068-2072.
- Sheng QH, Li JC, Alam MS, Fang XP, Guo MR. Gross composition and nutrient profiles of Chinese yak (Maiwa) milk. Int J Food Sci Technol 2008; 43: 568-572.
- Li HM, MaY, Li QM, Wang JQ, Cheng JJ, Xue J, Shi J. The chemical composition and nitrogen distribution of Chinese yak (Maiwa) milk. Int J Mol Sci 2011; 12: 4885-4895.
- Liu SC, Zhao X, Qian Y, Li J, Chen LH, Chen J, Suo HY. Identification of lactobacillus isolated from fermented yak milk in Tibetan Yangbajing area. Sci Technol Food Ind 2016; 37: 208-212.
- Jiang HY, Chen ZL, Zhao GH, Yang JX. Investigating the diversity of lactic acid bacteria in Tibetan traditional fermented dairy products by PCR-DGGE. Food Sci 2014; 35: 167-173.
- Iqbal MZ, Qadir MI, Hussain T, Janbaz KH, Khan YH, Ahmad B. Review: probiotics and their beneficial effects against various diseases. Pak J Pharm Sci 2014; 27: 405-415.
- Guandalini S. Probiotics for prevention and treatment of diarrhea. J Clin Gastroenterol 2011; 45: 149-153.
- Wu R, Wang L, Wang J, Li H, Menghe B, Wu J, Guo M, Zhang H. Isolation and preliminary probiotic selection of lactobacilli from koumiss in Inner Mongolia. J Basic Microbiol 2009; 49: 318-326.
- Khalil MA, El-Sheekh MM, El-Adawi HI, El-Deeb NM, Hussein MZ. Efficacy of microencapsulated lactic acid bacteria in Helicobater pylori eradication therapy. J Res Med Sci 2015; 20: 950-957.
- Li W, Ji J, Rui X, Yu JJ, Tang WZ, Chen XH, Jiang M, Dong MS. Production of exopolysaccharides by Lactobacillus helveticus MB2-1 and its functional characteristics in vitro. LWT-Food Sci Technol 2014; 59: 732-739.
- Wang J, Gu YJ, Ma WH, Yang ZN. Screening of potential probiotic lactic acid bacteria from Inner Mongolian dairy Tofu. Food Sci 2014; 35: 171-177.
- Chen XY, Li J, Zhao X, Qian Y, Chen LH, Chen J, Suo HY. In vitro screening of lactic acid bacteria with probiotic properties from traditional fermented Yak yogurt. Food Fermentat Ind 2015; 42: 85-90.
- Dong XW, Li BK, Li KX, Lu SL, Guo SJ, Peng XL. Diversified comparison of lactic acid bacteria from traditional dairy products in Mongolian and Kazakh in Xinjiang. Sci Technol Food Ind 2013; 34: 162-166.
- Yu J, Sun Z, Liu W, Zhang J, Sun T, Bao Q, Zhang H. Rapid identification of lactic acid bacteria isolated from home-made fermented milk in Tibet. J Gen Appl Microbiol 2009; 55: 181-190.
- Patel A, Prajapati JB, HolstO, Ljungh A. Determining probiotic potential of exopolysaccharide producing lactic acid bacteria isolated from vegetables and traditional Indian fermented food products. Food Biosci 2014; 5: 27-33.
- Tuo YF, Zhang WQ, Zhang LW, Ai LZ. Aessessment of the probiotc properties of lactobacillus strains. Mod Food Sci Technol 2012; 8: 906-910.
- Zhang HP, Menghebilige, Wang JG, Sun TS, Xu J, Wang LP, Yun YY, Wu RN. Assessment of potential probiotic properties of L.casei Zhang strain isolated from traditionally home-made Koumiss in Inner Mongolia of China. China Dairy Ind 2006; 4: 4-10.
- Chen X, Wu RN, Meng H, Zhang HP. Review on the mechanism of acid tolerance of lactic acid bacteria. China Dairy Ind 2008; 3: 30-34.
- Pan XD, Chen FQ, Wu TX. The acid, bile tolerance and antimicrobial property of Lactobacillus acidophilus NIT. Food Control 2009; 20: 598-602.
- Begley M, Gahan CGM, hill C. The interaction between bacteria and bile. FEMS Microbiol Rev 2005; 29: 625-651.
- Wang X, Hou JM, Fu LL, Yang SD, Zhang X, Li D, Zhang ZN. In vitro screening of lactic acid bacteria producing β-galactosidase and their probiotic characteristics. China Dairy Ind 2012; 40: 20-23.