ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 9
Investigation of antioxidant enzyme polymorphism in chronic obstructive pulmonary disease patients
Abdullah Arpaci1, Serap Yalin2, Devrim Taskin3, Fusun Fakioglu3, Kemal Turker Ulutas4* and Ulku Comelekoglu5
1Department of Medical Biochemistry, Ad?yaman University Faculty of Medicine, Ad?yaman, Turkey
2Department of Biochemistry, Mersin University, Mersin, Turkey
3Department of Chest Diseases, Ad?yaman Education-Research Hospital, Ad?yaman, Turkey
4Department of Medical Biochemistry, Mustafa Kemal University, Hatay, Turkey
5Department of Biostatistics, Mersin University, Mersin, Turkey
- *Corresponding Author:
- Kemal Turker Ulutas
Department of Medical Biochemistry
Mustafa Kemal University, Turkey
Accepted on March 7, 2018
DOI: 10.4066/biomedicalresearch.29-17-3854
Visit for more related articles at Biomedical ResearchBackground: Antioxidant system enzyme activity and polymorphism were investigated in Chronic Obstructive Pulmonary Disease (COPD), which is increasingly prevalent both in the world and our country.
Methods: Malondialdehyde (MDA) was measured to determine lipid peroxidation, while Catalase (CAT), Paraoxonase (PON) and Superoxide Dismutase (SOD) enzyme activities were measured to determine antioxidant activity. For molecular analysis, leukocytes were separated and DNA was isolated from cell nuclei and the targeted genes were amplified by using PCR. Targeted base changes were detected by the restriction fragment length polymorphism technique.
Results: Evaluating CAT polymorphism, there was 35 (35%) TT, 52 (52%) TC and 13 (13%) CC genotypes in the control group, as 38 (38%) TT, 40 (40%) TC and 22 (22%) CC genotypes in the patient. In SOD, there were 32 (32%) TT, 49 (49%) TC, 19 (19%) CC genotypes in the control group, as 34 (34%) TT, 50 (50%) TC and 16 (16%) CC genotypes in the patient. In PON55, there were 50 (50%) LL, 38 (38%) LM, 12 (12%) MM genotypes in the control group, as 39 (39%) LL, 49 (49%) LM and 12 (12%) MM genotypes in the patient. In PON192, there were 77 (77%) QQ, 21 (21%) QR, 2 (2%) RR genotypes in the control group, as 66 (66%) QQ, 29 (49%) QR, 5 (5%) RR genotypes in the patient. There was no significant difference in genotype distribution for CAT, SOD, PON55 and PON192 polymorphisms between groups.
Conclusion: In conclusion, COPD patients were found to have a deficiency in the antioxidant defense system. This will lead to the development of oxidative stress in patients and the complications of COPD. There was no polymorphic difference between the control group and South African patients living in Adiyaman in COPD whose genetic characteristics were not fully explained.
Keywords
Antioxidant, Enzyme polymorphism, Chronic obstructive pulmonary disease.
Introduction
Chronic Obstructive Pulmonary Disease (COPD) is a disease of the lungs caused by abnormal inflammatory response to harmful particles or gases, but also effects of systemic infection are also observed. According to this definition, COPD has an inflammation in the airways and lung parenchyma [1].
Findings which indicate that oxidant stress plays an important role in the pathogenesis of COPD increase. Lungs are more susceptible to oxidants as they are the most oxygenchallenging organs in the body. The classical oxidants that play a role in the development of COPD are superoxide (O2-), hydrogen peroxide (H2O2), hydroxyl (OH), peroxynitrite (ONOO) and isoprostanes. Oxidants contribute to the development of COPD by causing damage to protein, lipid, and nucleic acid structure, cell dysfunction, or death [1,2]. Smoking is also the most important cause of oxidative stress in COPD. Catalase, Superoxide Dismutase (SOD), glutathione, vitamins A and C, uric acid and lactoferrin are the most important antioxidants [3]. Free radicals cause oxidant/ antioxidant imbalance and cause damage to the epithelial cells in ventilation areas [4]. MDA is one of the most important lipid peroxidation products showing oxidative stress [5].
The development of COPD in only a proportion of smokers suggests genetic predisposition in the pathogenesis of the disease. The presence of a significant familial risk for airway stiffness in the smoking siblings of severe COPD patients also suggests that genetic predisposition is frequent [6]. Apart from smoking, there are other risk factors such as outdoor pollution, occupational dust exposure, high salt diet, dietary antioxidant deficiency, oxidants, and low birth weight [7]. Nonetheless, the development of COPD in one of the two subjects exposed at the same level is associated with the genetic predisposition of the person or how long they have lived (2). The World Health Organization (WHO) reported in 2011 as the 4th disease among the highest number of deaths among all age-related diseases with a rate of 5.1% [8]. In the United States, only the incidence of COPD has increased among the diseases that caused death in the last 30-40 years. COPD worldwide is estimated to be in the fifth position in 2030 when the disease burden is on the 13th place in 2004 [9].
In our study; we aimed to investigate the relationship between oxidant/antioxidant imbalance, antioxidant enzyme activities and genetic variations in this disease, which is an increasingly important health problem both in the world and in Turkey. For this purpose, we examined Malondialdehyde (MDA), an indicator of oxidative damage, and enzyme activities and gene polymorphisms of Paraoxanase (PON), catalase and SOD from antioxidant enzymes in patients admitted to Ad?yaman University Training-Research Hospital.
Material and Method
100 patients with COPD who were admitted to the Adiyaman State Hospital Internal Medicine Policlinic and 100 healthy subjects without COPD or any family history of COPD were included in the study. The study was approved by Ad?yaman University Clinical Research Ethics Committee. Both the patient and the control group were informed about the study before including in the study.
Analysis methods used for MDA level and enzyme activities
In blood samples taken from the cases, MDA levels were observed to determine lipid peroxidation and catalase, PON and SOD activities were observed to determine antioxidant levels. Venous blood samples from EDTA tubes were centrifuged for 10 min at 3000 rpm to separate the plasmas and stored at -20°C until analysis.
1. MDA Measurement; is based on the spectrophotometric evaluation of the absorbance of the pink red color resulting from the reaction of the MDA with thiobarbutyric acid, which is the most stable of the lipid peroxidation products [10].
2. Catalase enzyme activity assay; Catalase activity was determined according to the method described by Aebi. The method is based on the enzymatic degradation of the H2O2 substrate by catalase at 240 nm [11].
3. PON enzyme activity assay; PON activity assay was performed according to the method developed by Eckerson et al. PON activity was examined by measuring the formation of p-nitrophenol at 412 nm resulting from the enzymatic hydrolysis of PON by using 100 mM Tris-HCl, pH 8 buffer containing 2 mM CaCl2 and 4 mM paraoxon in the activity measurement [12].
4. SOD enzyme activity assay; principle of experiment: The measurement principle of SOD enzyme activity accelerating the aquatic and molecular oxygen dismutation of endogenous and exogenous sources of toxic superoxide radicals generated during the production of energy by oxidative way is based on measuring the color absorbed at 560 nm with Nitroblue Tetrazolium (NBT) superoxide radicals which xanthine oxidase releases into the presence of xanthine [13].
Molecular analysis
In the first step of the molecular analysis part of this study, leukocytes were isolated from the cell nuclei by separating the whole blood. DNA samples were stored at -20°C until analysis. In the second step, the targeted genes were amplified by PCR. The replicate samples were run on an agarose gel stained with ethidium bromide (EtBr2) and visualized with ultraviolet light. In the third step, targeted base changes were detected by Restriction Fragment Length Polymorphism (RFLP) technique.
PON 192 polymorphism analyses
In genomic DNA samples, the alleles of the PON 192 locus were amplified by Polymerase Chain Reaction (PCR). Primer sequences:
5’-TAT TGT TGC TGT GGG ACC TGA G-3’
5’-CCTGAGAATCTGAGTAAATCCACT-3’
The PCR products were cut with AlwI restriction endonuclease at 37°C for 8 h and analysed and genotyped on ethidium bromide stained 2% agarose gel electrophoresis. According to the genotype, the Q allele yields a single band at 238 bp, R allele two bands at 172 bp and 66 bp [14].
PON 55 polymorphism analyses
Alleles belonging to PON 55 locus were multiplied by PCR in genomic DNA samples of individuals. Primer sequences:
5’-CCTGCAATAATATGAAACAACCTG-3’
5’-TGAAAGACTTAAACTGCCAGTC-3’
The fragment of the PON 55 locus amplified by PCR was cut with NlaIII restriction endonuclease at 37°C for 2 h and then subjected to 2% agarose gel electrophoresis. DNA fragments were stained with ethidium bromide and then visualized under UV and genotyped. L aliquot yields a band at 172 bp, M allele two bands at 106 and 66 bp [15].
Catalase polymorphism analysis
Primer sequences used to identify catalase-262 C/T polymorphism:
F: 5’-TAAGAGCTGAGAAAGCATAGCT- 3’
R: 5-’AGAGCCTCGCCCCGCCGGACCG-3’
Amplification was performed and PCR products were cut with Sma I restriction endonuclease at 37°C for 16 h and then subjected to 2% agarose gel electrophoresis. DNA fragments were stained with ethidium bromide and then visualized under UV and genotyped. The T allele gives a band at 340 bp, Callele two bands at 185 and the 155 bp [16].
SOD polymorphism analysis; Primer sequences used to identify SOD gene Val-9Ala polymorphism was multiplied by:
F: 5’-ACC AAC CAG CAG GCA GCT GGC GCC GG-3’
R: 5’-GCG TTG ATG TGA GGT TCC AG-3’
And PCR products were cut with NgoM IV restriction endonuclease for 16 h at 37°C and then subjected to 2% agarose gel electrophoresis. DNA fragments were stained with ethidium bromide and then visualized under UV and genotyped. T-allele gives single band at 107 bp, C allele two bands at 89 and 18 bp. The 18 bp band could not be observed because the agarose gel was too small to appear on the gel and the evaluation was made accordingly [17].
Statistical analysis
Control was used for SOD, MDA PON, and catalase values, LSD multiple comparison tests were used for ANOVA and in subgroup comparisons to test statistical differences between COPD groups. The mean ± standard deviation values were given as descriptive statistics and Error bar graphs were drawn for the variables. Chi square and Fisher test were used to assess the frequency of occurrence of genotype and alleles among groups. Analyses were evaluated with MedCalc v.12.3 package program.
Results
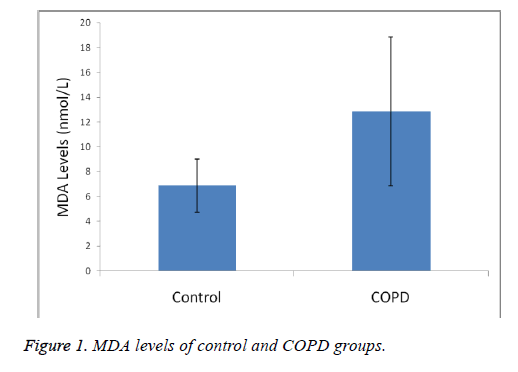
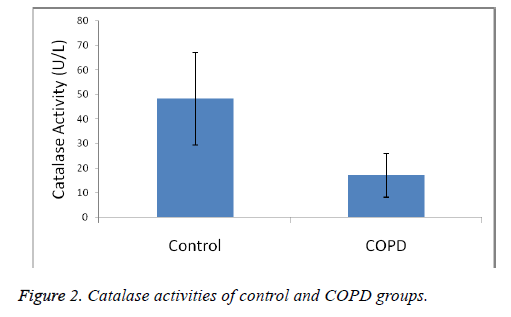
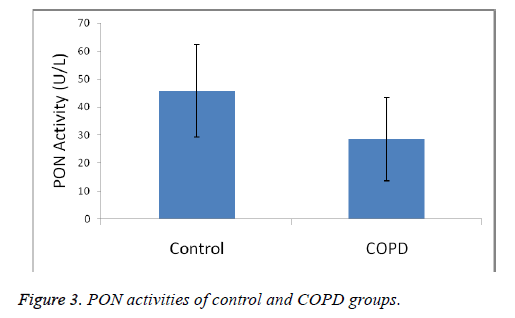
Plasma MDA levels and catalase, PON and SOD enzyme activities of all groups were shown in Figures 1-4. According to the findings obtained, MDA levels were found to be 6.88 ± 2.14 nmol/L in the control group and 12.88 ± 6.01 nmol/L in COPD patients. Plasma catalase levels were determined as 48.23 ± 18.86 U/L in the control group and 17.09 ± 8.82 U/L in COPD patients. Plasma PON activities were determined as 45.89 ± 16.46 U/L in the control group and 28.52 ± 14.85 U/L in COPD patients.
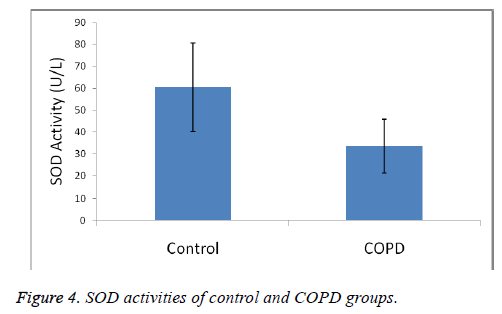
Plasma SOD activities were 60.36 ± 20.29 U/L in the control group and 33.44 ± 12.32 U/L in COPD patients.
A statistically significant increase in plasma MDA concentration (p<0.01) and a significant decrease in plasma catalase, PON and SOD activities (p<0.01) were observed when the data of COPD cases were compared with the data of the control group.
When the genotype findings of individuals with COPD and control group participating in the study were evaluated, it was found out that in the control group 35 individuals (35%) had TT genotype, 52 individuals (52%) had TC genotype, 13 individuals (13%) had CC genotype and in the patient group 38 individuals (38%) had TT genotype, 40 individuals (40%) had TC genotype and 22 individuals (22%) had CC genotype. There was no significant difference in genotype distribution in terms of catalase polymorphism between the groups (p>0.05) (Table 1).
| Catalase | Control (n=100) | COPD (n=100) | Chi-square | OR | |||
|---|---|---|---|---|---|---|---|
| Genotype | n | % | N | % | P | OR (95% GA) | P |
| TT | 35 | 35 | 38 | 38 | 0.135 | 1.00 | Reference |
| TC | 52 | 52 | 40 | 40 | 0.708 (0.38-1.31) | 0.274 | |
| CC | 13 | 13 | 22 | 22 | 1.559 (0.68-3.56) | 0.292 | |
| Allele frequency | n | % | N | % | |||
| T | 122 | 61 | 116 | 58 | 0.611 | - | |
| C | 78 | 39 | 84 | 42 | 1.133 (0.76-1.69) | 0.541 | |
Table 1. Distribution of catalase polymorphism genotype and allele frequencies in the control and COPD.
When the distribution of catalase polymorphism alleles was examined, the frequency of T allele was found to be 61% and the frequency of C allele was 39% in the control group, while the frequency of T allele and C allele were found to be 58% and 42% in the patient group, respectively (Table 1). There was no significant difference in allele frequencies (p>0.05).
When the genotype findings of individuals with COPD and control group participating in the study were evaluated, it was found out that in the control group, 32 individuals (32%) had TT genotype, 49 (49%) individuals had TC genotype, 19 individuals (19%) has CC genotype and in the patient group 34 individuals (34%) has TT genotype, 50 individuals (50%) had TC genotype, and 16 individuals (16%) had CC genotype. There was no significant difference between the groups in terms of genotype distribution for SOD polymorphism (Table 2).
| SOD | Control (n=100) | COPD (n=100) | Chi-square | OR | |||
|---|---|---|---|---|---|---|---|
| Genotype | n | % | N | % | P | OR (95% GA) | P |
| TT | 32 | 32 | 34 | 34 | 0.849 | 1.00 | Reference |
| TC | 49 | 49 | 50 | 50 | 0.960 (0.51-1.79) | 0.899 | |
| CC | 19 | 19 | 16 | 16 | 0.793 (0.35-1.80) | 0.579 | |
| Allele frequency | n | % | N | % | |||
| T | 113 | 56.5 | 118 | 59 | 0.686 | - | |
| C | 87 | 43.5 | 82 | 41 | 0.903 (0.61-1.34) | 0.613 | |
Table 2. Distribution of SOD polymorphism genotype and allele frequencies in control and COPD groups.
When the distribution of SOD polymorphism alleles was examined, the frequency of T allele was found to be 56.5% and the frequency of C allele was 43.5% in the control group while the frequency of T allele and C allele were found to be 59% and 41% in the patient group (Table 2). There was no significant difference in allele frequencies.
When the genotype findings of PON55 polymorphism of the subjects were evaluated in the COPD and control groups participated in the study, it was found out that 50 individuals (50%) has LL genotype, 38 individuals (38%) had LM genotype, 12 individuals (12%) had MM genotype, and in the patient group 39 individuals (39%) had LL genotype, 49 individuals (49%) had LM genotype and 12 individuals (12%) had MM genotype. There was no significant difference in genotype distribution in terms of PON55 polymorphism between groups (Table 3).
| PON | Control (n=100) | KOAH (n=100) | Chi-square | OR | |||
|---|---|---|---|---|---|---|---|
| Genotype | n | % | N | % | P | OR (95% GA) | P |
| LL | 50 | 50 | 39 | 39 | 0.253 | - | |
| LM | 38 | 38 | 49 | 49 | 1.653 (0.91-2.99) | 0.098 | |
| MM | 12 | 12 | 12 | 12 | 1.282 (0.52-3.16) | 0.589 | |
| HW (p) | |||||||
| L | 138 | 69 | 127 | 63.5 | 0.290 | --- | |
| M | 62 | 31 | 73 | 36.5 | 1.279 (0.84-1.94) | 0.245 | |
Table 3. Distribution of PON55 polymorphism genotype and allele frequencies in the control and COPD.
When the distribution of PON55 polymorphism alleles was examined, the frequency of L allele was found 69% and the frequency of M allele was 31% in the control group, while the frequency of L allele was 63.5% and M allele frequency was 36.5% in the patient group (Table 3). There was no significant difference in allele frequencies.
When the genotype findings of PON192 polymorphism of individuals from the study participating COPD and control group were evaluated, it was found out in the control group that 77 individuals (77%) has QQ genotype, 21 individuals (21%) had QR genotype, 2 individuals (2%) had RR genotype and in patient group 66 individuals (66%) had QQ genotype, 29 individuals (29%) had QR genotype, and 5 individuals (5%) had the RR genotype. There was no significant difference in genotype distribution in terms of PON192 polymorphism between the groups (p>0.05) (Table 4).
| PON | Control (n=100) | COPD (n=100) | Chi-square | OR | |||
|---|---|---|---|---|---|---|---|
| Genotype | n | % | N | % | P | OR (95% GA) | P |
| 77 | 77 | 66 | 66 | 0.179 | - | ||
| QR | 21 | 21 | 29 | 29 | 0.623 (0.33-1.19) | 0.151 | |
| RR | 2 | 2 | 5 | 5 | 0.342 (0.06-1.83) | 0.257 | |
| Allele frequency | n | % | N | % | |||
| Q | 175 | 88 | 161 | 81 | 0.057 | - | |
| R | 25 | 12 | 39 | 19 | 0.591 (0.34-1.02) | 0.057 | |
Table 4. Distribution of PON192 polymorphism genotype and allele frequencies of control and COPD.
When the distribution of PON192 polymorphism allele frequencies were examined, the frequency of Q alleles was found to be 88% and the frequency of R alleles 12% in the control group, while the frequency of Q alleles was found 81% and M allele frequencies were found 19% in the patient group (Table 4). There was no significant difference in allele frequencies.
Discussion
COPD is a growing public health problem. 80 million people have been affected moderately or severely, and it is estimated that in 2030, it will rise to the 3rd place among the causes of death [8]. When COPD is considered to be a male disease, the number of women who died of COPD in the United States in 2000 was traced to males and there was no difference in terms of gender. In underdeveloped and developing countries such as Turkey, the air pollution due to the use of wood and biomass fuel and the increase in the elderly population can be considered to raise the COPD burden [4,18].
The World Health Organization (WHO) reported in 2011 that it was at 4th rank among all age-related diseases which cause the highest number of deaths with a rate of 5.1%. In the United States, only the incidence of COPD has increased in the past 30-40 years among the diseases leading to death [8]. In addition to mechanisms such as proteolytic-oxidative damage, endothelial dysfunction; genetic, diet and infection heterogeneity are also considered as other factors in the pathogenesis of COPD [19]. Smoking is a predominant cause of COPD with a rate of 80-90%. However, COPD's clinical signs are seen in 15-20% of smokers [20,21]. Oxidative stress plays an important role in pathogenesis of certain diseases such as lung cancer, COPD, arteriosclerosis [22]. This results in neutrophil infiltration and impaired ocular/antioxidant balance [23,24].
MDA levels, an end product of lipid peroxidation and indicative of oxidative stress, were significantly increased in COPD patients, as was also the case in other studies (p<0.01). Catalase, SOD and PON plasma activities have found as decreased. Our results shown in Figures 1-4 show that oxidant/ antioxidant balance is impaired in our COPD patients (p<0.01) [25-30].
Several candidate genes have been reported that are significantly related to COPD phenotypes (1). Both oxygenase 1, SOD, catalase, GST, microsomal epoxide hydrolase (EPHX1) and cytochrome P450 are antioxidant and detoxifying enzymes, which play a protective role in the respiratory tract against oxidative damage [31]. Functional genetic variations may therefore be important in altering the antioxidant capacity of the lung. Superoxide Dismutase (SOD), Catalase (CAT), Glutathione peroxidase (GSH-px), Glutathione Reductase (GR), Glutathione-S transferase P1 (GSTP1) and several oxidative detoxification and antioxidant gene polymorphisms such as Oxygenase-1 (HO-1), Microsomal epoxy hydrolase-1 (EPHX1) were also investigated with suspicion of COPD [17,32]. EC-SOD, SOD3 were significantly associated with COPD, and conflicting results were obtained with others, the role of genetic diversity in COPD is not yet well understood and COPD is offered as a complex disease characterized by polygenic nature due to random activation of several genetic events. Recent reports have suggested the diversity of polymorphisms of genes coding for increased frequency of detoxification enzymes among COPD patients with different ethnicity and circles [14,33-36].
As is known, Paraoxonase (PON1) is a calcium-dependent ester hydrolase with High Density Lipoproteins (HDL) and a protein consisting of 354 amino acids with a molecular weight of 43 kDa [37]. It is synthesized in the liver. Parathion, an organophosphorous insecticide, has the property of hydrolyzing paraoxon active metabolite. PON has two main functions and its enzymatic activity shows individual differences [14]. PON1 alveolar epithelium plays a role in protecting the type1 cells, endothelial cells and Clara cells from localized and pulmonary oxidative stress. Smoking causes airborne epithelial damage, resulting in oxidative stress, and immediately there is a decrease in PON1 levels [22]. Cigarette smoke extracts are known to cause inhibition by modifying the free thiol groups of PON enzyme [21]. Although PON1 has been extensively studied in diabetes and cardiovascular diseases [23], few have been studied in COPD. We studied 192 and 55 polymorphisms of PON in our study but could not find any significant difference from the control group.
Extracellular SOD (SOD3) is the primary extracellular antioxidant enzyme in the lungs and has been shown to protect the extracellular matrix during lung injury [24]. However, it has been suggested that genetic variants in SOD3 may reduce pulmonary function and lead to COPD in adults [25]. In the studies of Juul et al. [26] in 9258 Danish individuals and Young et al. [31] in 440 individuals from European general population reached the conclusion that found that the ECSOD R213G mutation protects smokers from COPD. Dahl et al. have reported that ECSOD E1/I1 polymorphism is not associated with COPD in the study of 35635 people in Copenhagen general population, whereas in Copenhagen city heart study which was conducted on 9093 subjects and mentioned that ECSOD E1/I1 polymorphism was associated with COPD morbidity [27]. We studied MnSOD Val-9Ala polymorphism and found no statistical significance between COPD and control group. This issue is open to debate and needs to be studied.
Catalase gene is localized in chromosome 11p13 and consists of 13 exons. Several polymorphisms have been reported that decrease the erythrocyte catalase level of this enzyme-262 C/T polymorphism is associated with basal catalase expression and erythrocyte levels [11,28]. Mak et al. have studied iSOD Ala16Val and -262 C/T functional catalase polymorphism in Chinese society. They found no difference between the control group and COPD [33]. Young et al. studied catalase polymorphism, COPD and resistance in smokers and found no significant allelic difference, suggesting that more studies should be conducted for the relation of antioxidant genes with by comparing the control group in the same ethnic group and geographical regions [31].
In a study of Aynacioglu et al. conducted in our country, a negative association between COPD and Arg192Gln polymorphism was found [29]. In this study, the relationship between COPD and PON1 Leu55Met and Gln192Arg polymorphisms was examined. PON1, the gene encoding PON1 enzyme, mainly contains two important polymorphisms. These include a polymorphism of 55 L/M with methionine (Met-M) instead of leucine (Leu-L) at position 55 and 192 Q/R polymorphism where include Arginine (Arg-R) instead of Glutamine (Gln-Q) at position 192. In 192 Q/R-polymorphism, it is observed that those with Gln alleles have a lower PON1 enzyme activity than those with the Arg allele [30]. In 55 L/M polymorphism, MM homozygous individuals have lower PON1 activity against paraoxon compared to LL homozygotes [29].
In conclusion, we investigated the activities of superoxide dismutase, catalase and paroxanase enzymes, which are markers of lipid peroxidation in patients with COPD, who struggle with MDA and oxidant stress. In line with the literature, we found a rise in MDA and a decrease in antioxidant enzyme activities. We also tried to look at the genetic polymorphisms of these enzymes. However, we did not find any significant difference with the control group. In the literature, genetic studies have both inadequate and contradictory results. Different ethnic groups have different outcomes. We believe that more detailed studies are necessary.
Acknowledgements
This work was supported by Ad?yaman University Scientific Research Projects Coordinator.
References
- Barnes PJ, Shapiro SD, Pauwels RA. Chronic obstructive pulmonary disease: molecular and cellularmechanisms. Eur Resp J 2003; 22: 672-688.
- Chung KF, Adcock IM. Multifaceted mechanisms in COPD: inflammation, immunity, and tissue repair and destruction. Eur Resp J 2008; 31: 1334-1356.
- Ulutas KT, Buyukbas S, Yilmaz N, Arpaci A, Kaya H. Association of fetuin-a with glucose-6-phosphate dehydrogenase and oxidative stress in sickle cell anemia. Acta Medica Mediterranea 2016; 32: 959-966.
- MacNee W. Oxidants/antioxidants and COPD. Chest 2000; 117: 303S-17S.
- Uzun K. Oxidative stress in smokers and COPD. T Klin J Med Sci 1999; 19: 123-129.
- Agustí AGN, Noguera A, Sauleda J, Sala E, Pons J, Busquets X. Systemic effects of chronic obstructive pulmonary disease. Eur Resp J 2003; 21: 347-360.
- Bates D, Breese L, brocklebank C, zarnke k, Jamieson P, Mackay E. Venous thromboembolism prophylaxis in obese medical patients: a retrospective cohort study. Ulutas Med J 2015; 1: 104-110.
- World Health Organization (WHO) WHO. Chronic respiratory diseases. WHO. http: //www.who.int/ respiratory/copd/burden/en/index.html2011.
- Gunen H, Hacievliyagil SS, Yetkin O, Gulbas G, Mutlu LC, Pehlivan E. Prevalence of COPD: first epidemiological study of a large region in Turkey. Eur J Int Med 2008; 19: 499-504.
- Yagi K. Simple procedure for specific assay of lipid hydroperoxides in serum or plasma. Methods Mol Biol 1998; 108: 107-110.
- Aebi H. Catalase in vitro. Methods Enzymol 1984; 105: 121-126.
- Eckerson HW, Wyte CM, La Du BN. The human serum paraoxonase/arylesterase polymorphism. Am J Human Gene 1983; 35: 1126-1138.
- Sun Y, Oberley LW, Li Y. A simple method for clinical assay of superoxide dismutase. Clin Chem 1988; 34: 497-500.
- Lakshmy R, Ahmad D, Abraham RA, Sharma M, Vemparala K, Das S. Paraoxonase gene Q192R and L55M polymorphisms in Indians with acute myocardial infarction and association with oxidized low density lipoprotein. Ind J Med Res 2010; 131: 522-529.
- Humbert R, Adler DA, Disteche CM, Hassett C, Omiecinski CJ, Furlong CE. The molecular basis of the human serum paraoxonase activity polymorphism. Nat Gene 1993; 3: 73-76.
- Goulas A, Fidani L, Kotsis A, Mirtsou V, Petersen RC, Tangalos E. An association study of a functional catalase gene polymorphism, -262C → T, and patients with Alzheimers disease. Neurosci Lett 2002; 330: 210-212.
- Cai Q, Shu XO, Wen W, Cheng JR, Dai Q, Gao YT. Genetic polymorphism in the manganese superoxide dismutase gene, antioxidant intake, and breast cancer risk: results from the Shanghai Breast Cancer Study. Breast Cancer Res 2004; 6.
- Altuntaay E, Turgut T, Ilhan N, Deveci F, Muz HM, Celik I. The levels of oxidant and antioxidant in patients with COPD. Tuberk Toraks 2003; 51: 373-379.
- Saryal SB, Acican T. Güncel bilgiler isiginda KOAH. Patogenez ve Inflamasyon 2003; 22: 21-33.
- Nishio E, Watanabe Y. Cigarette smoke extract inhibits plasma paraoxonase activity by modification of the enzymes free thiols. Biochem Biophys Res Commun 1997; 236: 289-293.
- Isik B, Ceylan A, Isik R. Oxidative stress in smokers and non-smokers. Inhal Toxicol 2007; 19: 767-769.
- Durrington PN, Mackness B, Mackness MI. Paraoxonase and atherosclerosis. Arterioscler Thromb Vasc Biol 2001; 21: 473-480.
- Pinar T, Cevat S, Nurullah T. The relationship between paraoxanase gene Leu-Met (55) and Gln-Arg (192) polymorphisms and coronary artery disease. Arch Turk Soc Cardiol 2009; 37: 473-478.
- Fattman CL, Schaefer LM, Oury TD. Extracellular superoxide dismutase in biology and medicine. Free Radic Biol Med 2003; 35: 236-256.
- Oberley-Deegan RE, Regan EA, Kinnula VL, Crapo JD. Extracellular superoxide dismutase and risk of COPD. COPD J Chron Obstruct Pulm Dis 2009; 6: 307-312.
- Juul K, Tybjærg-Hansen A, Marklund S, Lange P, Nordestgaard BG. Genetically increased antioxidative protection and decreased chronic obstructive pulmonary disease. Am J Resp Crit Care Med 2006; 173: 858-864.
- Dahl M, Bowler RP, Juul K, Crapo JD, Levy S, Nordestgaard BG. Superoxide dismutase 3 polymorphism associated with reduced lung function in two large populations. Am J Resp Crit Care Med 2008; 178: 906-912.
- Quan F, Korneluk RG, Tropak MB, Gravel RA. Isolation and characterization of the human catalase gene. Nucleic Acids Res 1986; 14: 5321-5335.
- Aynacioglu AS, Kepekci Y. The human paraoxonase Gln-Arg192 (Q/R) polymorphism in Turkish patients with coronary artery disease. Int J Cardiol 2000; 74: 33-37.
- Adkins S, Gan KN, Mody M, La Du BN. Molecular basis for the polymorphic forms of human serum paraoxonase/arylesterase: glutamine or arginine at position 191, for the respective A or B allozymes. Am J Hum Genet 1993; 52: 598-608.
- Young R, Hopkins R, Black P, Nel Nee Eddy C, Wu L, Gamble G. Functional variants of antioxidant genes in smokers with COPD and in those with normal lung function. Resp Med COPD Update 2006; 1: 141.
- Tavilani H, Nadi E, Karimi J, Goodarzi MT. Oxidative stress in COPD patients, smokers and non-smokers subjects. Resp Care 2012.
- Mak JCW, Ho SP, Yu WC, Choo KL, Chu CM, Yew WW. Polymorphisms and functional activity in superoxide dismutase and catalase genes in smokers with COPD. Eur Resp J 2007; 30: 684-690.
- Tan RJ, Fattman CL, Watkins SC, Oury TD. Redistribution of pulmonary EC-SOD after exposure to asbestos. J Appl Physiol (1985) 2004; 97: 2006-2013.
- Bowler RP, Barnes PJ, Crapo JD. The role of oxidative stress in chronic obstructive pulmonary disease. COPD J Chron Obstruct Pulm Dis 2004; 1: 255-277.
- Lakhdar R, Denden S, Kassab A, Leban N, Knani J, Lefranc G. Update in chronic obstructive pulmonary disease: role of antioxidant and metabolizing gene polymorphisms. Exp Lung Res 2011; 37: 364-375.
- James RW, Leviev I, Righetti A. Smoking is associated with reduced serum paraoxonase activity and concentration in patients with coronary artery disease. Circulation 2000; 101: 2252-2257.