ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Artificial Intelligent Techniques for Bio Medical Signal Processing: Edition-I
Integrative support vector machine for the prediction of zinc-binding sites in proteins
Zinc binding proteins play an important role in biological function, many researches focus on the area of zinc-binding sites. Taking into account the advantages of support vector machine, based on the different tools for the prediction of zinc-binding sites, a novel predictor named combZincPred was proposed to integrate these result scores. Tested on a non-redundant dataset, AURPC of our method increased more, and other indexes are also better than the other three predictors. The method can be better used to the inference of zinc-binding protein function.
Keywords
Zinc-binding sites, Support vector machine, Prediction, Integrative.
Introduction
Metal ions are rich in proteins thus far investigated [1]. Among different kinds of metal ions present in the life, zinc is the second abundant trace elements after iron. Zinc binding with protein has a wide variety of biological functions, such as catalysis, structural stability and regulatory roles. In particular, it contributes to the treatment of a number of medical diseases, as well as the development of some drugs [2]. By analysis of the human genome, there are up to 3000 proteins participating in binding with zinc [3]. Therefore, focus on the research of zinc-binding sites has important significance. Zinc-binding sites are defined as amino acid residues in a protein, in which the distance of any one of the nitrogen atoms and oxygen atoms or sulfur atoms and zinc ions within 3.0 Å. Cysteine (CYS), Histidine (HIS), Aspartate (ASP) and Glutamic (GLU) are predominant four binding amino acid residues.
The traditional method of recognizing zinc-binding sites is through biological experiments, which owns time-consuming and expensive cost. The identification of zinc-binding site cannot be from the whole proteome. Afterwards, some bioinformatics-based prediction has been developed for solving the problems. Passerini et al. [4] presented a two-stage machine-learning approach combining support vector machines and neural networks, which predicted HIS and CYS at 73% precision and 61% recall. Then, they presented a better method called zincFinder [5] based on support vector machines for the identification of zinc-binding sites with human proteins. Shu [6] introduced a novel method named zincPred by combining support vector machine and homology-based on a non-redundant protein data set, in which precision with an increase of 10%. Zheng et al. [7] integrated three kinds of protein features and proposed a two-step random forest algorithm called zinc Identifier. AUC and AURPC of the method were respectively better than other methods. Chen [8] stated a hybrid method called zincExplorer only based on sequence, it integrates the outputs of SVM-, cluster- and template-based predictors, which achieved an AURPC of 0.851. Based on the different selections of protein features, some other algorithms have been developed to predict the binding sites [9-11].
Recently, there are a lot of tools for the prediction of zinc-binding sites. All of them are constructed mainly from two aspects to improve the prediction accuracy. One is the integration of different protein features; the other is the choice or the integration of different classical algorithms. Less researchers focus on the study of existed prediction tools. In view of this, we plan to study the tools, and support vector machine is used to integrate three famous predictive tools.
Materials and Method
Data set
So far, there are no uniform experimental data sets for the prediction of zinc-binding sites. Here data set from Zhao [9] was selected as the experiment data. The overlapped proteins in the data set with the Passerini_dataset from Passerini [4] were deleted in order to avoid over fitting. Finally, 392 protein chains were adopted.
Integrated model
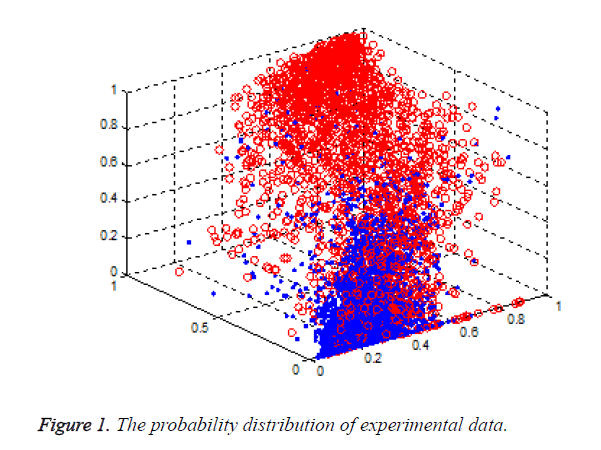
Support Vector Machine (SVM) is a new method of classification and pattern recognition, built on the basis of the principle of structural risk minimization, which has a strong learning ability and generalization performance. It shows superior performance, in solving small sample, nonlinear problem, and has been successfully applied in machine learning, data mining, and neural network. In this paper, we used the method to integrate the different tools for the prediction of the zinc binding sites. A novel method named combZincPred was presented. The scores of three famous predictors [5-7] were taken as the three features of the classification. As shown in Figure 1, it has a good discrimination for the two kinds of samples when using the three score features.
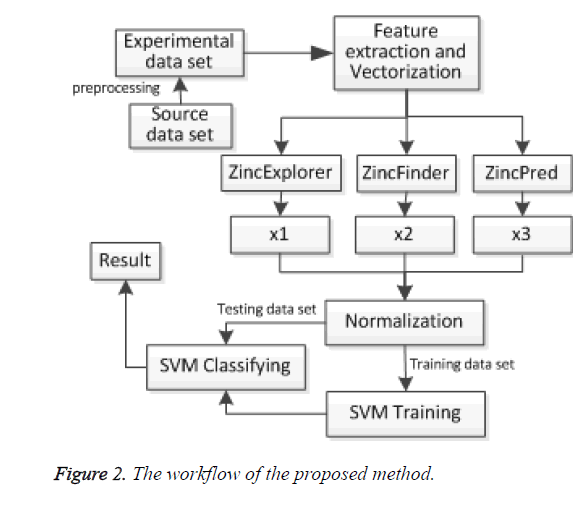
The scores x1, x2 and x3 of three predictors were normalized firstly. Then the experimental data was divided into training set and testing set. We obtained the support vector from training data set, and classified the test data set finally. The model framework is shown in Figure 2.
Performance evaluation
RPC (Recall-Precision Curve) is used to compare the performance of different predictors. The index AURPC (Area under RPC curve) is used to assess the predictor quantitatively. The value is between 0.5 and 1, the higher the value, the better the performance.
Results and Discussion
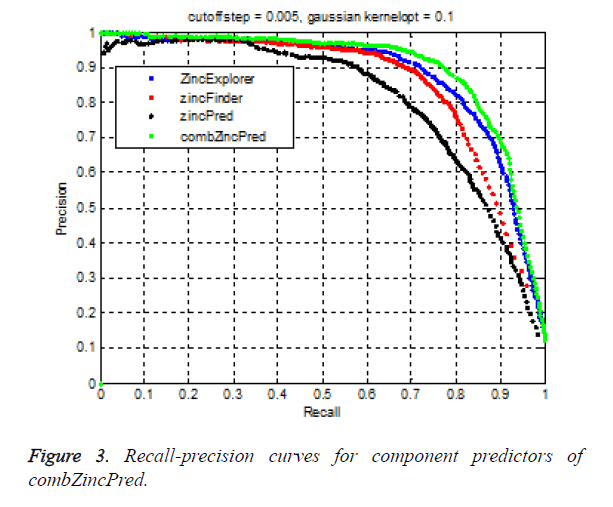
In the experiment, 10000 samples were chosen as the training set, and other 6000 samples were collected as the test set. After several rounds of testing, the Gauss kernel parameter is determined to be 0.1. Here cut off was selected as 0.005. Then we classify the test set using the trained model. RPC curves of four predictors were shown in Figure 3. RPC curve of combZincPred is above the curves of other three methods. AURPC of combZincPred reached nearly 0.88, in which Precision was 93.5%, specifity was 99.2% at a recall of 70%. AURPC of our method increased more than ZincExplorer, zincFinder and zincPred predictor at a recall of 70% respectively.
Conclusions
In order to predict the zinc-binding sites more accurately, an integrative predictor termed combZincPred was presented in the paper. Support vector machine is used to integrate the results of three famous prediction tools, considering its strong learning ability and generalization performance. Tested on a non-redundant dataset, AURPC of our method increased much more than the other three predictors and other indexes are also better. The method can be better used to the inference of zinc-binding protein function.
Acknowledgement
This work was supported by the Fundamental Research Funds for the Central Universities (NZ2013306), Higher School of Natural Science Research Project of Anhui Province (KJ2014A285), Natural Science Youth Fund of Anhui Province (1508085QF134), Top-notch Academic Programs Project of Jiangsu Higher Education Institutions, Supported by the Construct Program of Discipline in Software Engineering, Special research project of Humanities and social science of Ministry of Education (Engineering Science and technology personnel training).
References
- Ibers JA, Holm RH. Modeling coordination sites in metallobiomolecules. Science 1980; 209: 223-235.
- Chasapis CT, Loutsidou AC, Spiliopoulou CA, Stefanidou ME. Zinc and human health: an update. Arch Toxicol 2012; 86: 521-534.
- Kochanczyk T, Drozd A, Krezel A. Relationship between the architecture of zinc coordination and zinc binding affinity in proteins-insights into zinc regulation. Metallomics 2015;7: 244-257.
- Passerini A, Punta M, Ceroni A, Rost B, Frasconi P. Identifying cysteines and histidines in transition-metal-binding sites using support vector machines and neural networks. Proteins 2006; 65: 305-316.
- Passerini A, Andreini C, Menchetti S, Rosato A, Frasconi P. Predicting zinc binding at the proteome level. BMC Bioinformatics 2007; 8: 39.
- Shu N, Zhou T, Hovmoller S. Prediction of zinc-binding sites in proteins from sequence. Bioinformatics 2008; 24: 775-782.
- Zheng C, Wang M, Takemoto K, Akutsu T, Zhang Z. An integrative computational framework based on a two-step random forest algorithm improves prediction of zinc-binding sites in proteins. Plos One 2012; 7: 49716.
- Chen Z, Wang Y, Zhai YF, Song J, Zhang Z. ZincExplorer: an accurate hybrid method to improve the prediction of zinc-binding sites from protein sequences. Molecular Bio Sys 2013; 9: 2213-2222.
- Zhao W, Xu M, Liang Z, Ding B, Niu L. Structure-based de novo prediction of zinc-binding sites in proteins of unknown function. Bioinformatics 2011; 27: 1262-1268.
- He W, Liang Z, Teng M, Niu L. mFASD: a structure-based algorithm for discriminating different types of metal-binding sites. Bioinformatics 2015; 31: 1938-1944.
- Liu Z, Wang Y, Zhou C, Xue Y, Zhao W. Computationally characterizing and comprehensive analysis of zinc-binding sites in proteins. BiochimBiophysActa 2014; 1844: 171-180.