ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2010) Volume 21, Issue 2
Insilico docking for validation of drug leads on Superoxide dismutase of Homo sapiens and Plasmodium falciparum
1Department of Biochemistry, Yenepoya Medical College, Yenepoya University, Deralakatte, Mangalore 575018.
2Department of Microbiology, Yenepoya Medical College, Yenepoya University, Deralakatte, Mangalore 575018.
- *Corresponding Author:
- Vinitha R. Pai
Department of Biochemistry, Yenepoya Medical College
Yenepoya University, Deralakatte Mangalore 575018
Karnataka, India
E-mail: vinitharpai@gmail.com
Phone: +91 9980145182
Accepted date: October 07 2009
Superoxide dismutases (SODs) are metalloproteins which bring about dismutation of super-oxides and prevent biological damage of cellular components during oxidative stress. The malarial parasite plasmodium falciparum has a well developed defense system which in-cludes superoxide dismutase, to scavenge free radicals and combat oxidative stress during the erythrocytic stage of its life cycle. Alignment studies of the primary structure of plasmo-dium falciparum SOD (PfFe-SOD) have shown it to be structurally distinct from human cy-tosolic copper-zinc SOD1 (Cu-ZnSOD1). This feature makes it a potential target for ther-apy. A search for new targets as well as drugs is essential due to the phenomenon of drug re-sistance to chloroquine which is the drug of choice for treatment of malaria. A few potential inhibitors from the library of synthetic molecules have already been studied using recombi-nant PfFe-SOD by Soulere et al. Three of the fifteen effective lead molecules SP72, SP13 and SP59 show significant inhibition of recombinant PfFe-SOD. However, no attempt was made to investigate whether these lead molecules interact with the host Cu-ZnSOD1. In this study, the lead molecules showing significant inhibition were prepared using Marvin sketch. Cu-ZnSOD1 and PfFe-SOD was docked to these lead molecules and the energy values obtained. The results indicate almost equal affinity of the lead molecules to both, host and parasite, SOD. In conclusion, this study shows that compounds which are found to be quite effective inhibitors of recombinant enzyme in vitro need to be validated by alternate methods since, in the living system, compounds tend to deviate from their in vitro behavior in an unpredicted way. Computational tools such as insilico docking provide the scientist with an alternate base for validation of lead molecules.
Keywords
Plasmodium falciparum, Homo sapiens, insilico docking, chloroquine, Escherichia coli, computational bio-informatics tools.
Introduction
Generation of reactive oxygen species (ROS) results in oxidative stress which is responsible for the damage of cellular components [1]. Dismutation of superoxides can prevent such biological damage. Superoxide dismutases (SOD’s), catalases and glutathione peroxidases are some of the enzymes which catalyse dismutation of superoxide free radicals [2]. The malarial parasite Plasmodium falciparum is exposed to oxidative stress during the erythro-cytic stages of its life cycle [3]. The parasite has a well developed defense system with SOD and thioredoxin de-pendent peroxidases to scavenge free radicals generated in oxidative stress [4].
SODs are metalloproteins. Based on the metal cofactor that is associated with them SODs are classified as (i) Copper-Zinc SOD1 (Cu-ZnSOD1) exclusively found in eukaryotes (ii) Manganese dependent (Mn-SOD2) which is found in mitochondria and prokaryotes (iii) Iron de-pendent SOD (Fe-SOD) which is the most primitive form and associated with prokaryotes, protozoans, algae, an-aerobic bacteria and some higher plants [5]. Studies of primary structure alignment of these SODs have shown the Plasmodium falcifarum iron superoxide dismutase (PfFe-SOD) is structurally similar to Mn-SOD2 and Es-cherichia coli Fe-SOD but distinct from human Cu-ZnSOD1 [6]. This confirmed the suggestion that PfFe-SOD could be an ideal target for anti-malarials in the host erythrocytic phase of the parasite life cycle. Since eryth-rocytes do not have mitochondria and the related Mn-SOD2 isoform, the only interference is from the host Cu-ZnSOD1 which happens to be structurally distinct from the parasite PfFe-SOD. Therefore, inhibitors that are spe-cific for the PfFe-SOD could contribute as new drug leads against the parasite and will be a step towards develop-ment of advanced chemotherapy for malaria [7]. A few potential inhibitors from the library of synthetic mole-cules have already been studied using recombinant PfFe-SOD and the results were confirmed using two different SOD assays. Selection of possible leads was based on their structural diversity and their ability to create interac-tions of stacking, polar or hydrophobic nature. Two strains of Plasmodium falciparum, HB3 (which is sensi-tive to chloroquine, a first line drug of malaria) and Dd2 (which is moderately resistant to chloroquine) were used to test the in vitro efficacy of the most effective leads among those tested on the parasite enzyme. Among the fifteen effective lead molecules SP72, SP13 and SP59 showed significant inhibition of PfFeSOD. Since PfFeSOD is a unique enzyme, these lead molecules could have anti-malarial activity. The IC50 values were in μM range and it was suggested by the authors that this work could be the first step in the search for an anti-malarial for Plasmodium falciparum with PfFe-SOD as target. How-ever, no attempt was made in the study to check whether the molecules interact with and inhibit the host Cu-ZnSOD1 (from the erythrocyte cytosol). Our aim was to predict the ability of the prospective lead molecules to bind to both, the host Cu-ZnSOD1 and PfFe-SOD. Hence, it was of interest to try insilico docking of some of the lead molecules tested, on the 3D structures of SOD, both, from Homo sapiens (host) and the Plasmodium falcipa-rum (parasite). This study involves insilico docking of lead molecules on both, the host and parasite SOD. Lead molecules which effectively inhibited the recombinant PfFe-SOD were selected. The results are suggestive of affinity of the lead molecules to both SODs based on their energy values. If the lead molecules were to bind to host Cu-ZnSOD1 as well, it could lead to oxidative stress in the host which may be lethal. In conclusion, these results suggest that insilico docking of the lead molecules could give a clue as to whether they bind to the host Cu-ZnSOD1 with lesser energy than PfFe-SOD which is sug-gestive of an easier interaction with the former. Secondly, potential lead molecules may be used for an inhibition assay with recombinant Cu-ZnSOD1 as well, before be-ing tried as drugs against Plasmodium falciparum.
Materials and Methods
Computational bioinformatics tools have been employed in this study. Biological databases like PubMed and PDB (Protein Data Bank) and computational tools like Hex, Marvin Sketch and MolConvert have been used. PubMed is a digital archive of biomedical and life sciences journal literature managed by NCBI (National Center for Bio-technology Information) which serves as a search engine for accessing the MEDLINE database of citations and abstracts of biomedical research articles [8].
PDB (protein data bank) is a structural archive for bio-logical macromolecules which encompasses all the struc-tural information of macromolecules as determined by X-ray crystallography, NMR studies, etc. [9]. HEX is an interactive molecular graphics program for calculating and displaying feasible docking models of protein - ligand pairs of molecules which are based on HEX algorithm. It explores the space of all possible binding modes and iden-tifies the energy of particular molecular confirmation and then searches for a confirmation that minimizes the free energy of the system [10]. It uses spherical polar Fourier (SPF) correlation to accelerate the calculation and has inbuilt graphics to view the results [11].
Marvin Sketch is a chemical drawing tool which helps to quickly and easily draw molecules and chemical struc-tures [12].
Molconvert is a tool which further converts the files in .mol format of Marvin Sketch to .pdb format which be-comes accessible to hex algorithm [13].
Methodology
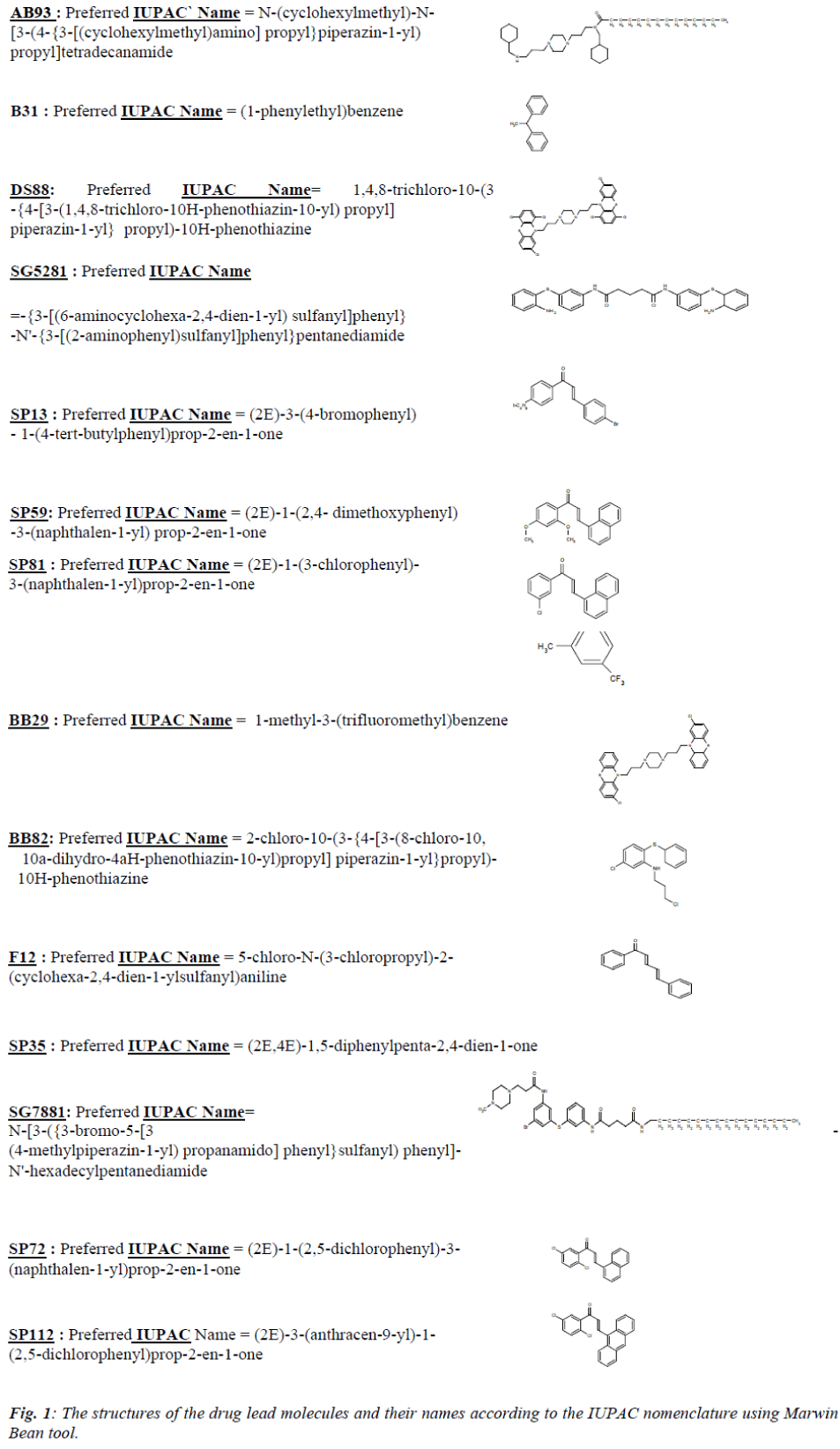
The 3D structures of the fourteen ligands from Soulere et al. [7] were drawn and the IUPAC names were assigned using the Marvin Sketch tool (Fig. 1). The structures gen-erated as a .mol file were converted to .pdb file using tool MolConvert which makes the lead molecule accessible to docking. The three-dimensional structure of PfFe-SOD and Cu-ZnSOD1 was retrieved from PDB. Taking PfFe-SOD, as a receptor, the docking analysis was done with the individual lead molecules as ligand using the docking software Hex 4.2. Similarly, docking was carried out with human Cu-ZnSOD1 as the receptor with the same lead molecules. The relative stabilities were evaluated using molecular dynamics and their binding affinity using free energy simulations. The receptors and the lead molecules were docked using the following parameters.
Display mode - polygons
Sort solutions by - energy
Correlation type - Shape only
FFT mode - 3D Fast lite
Grid dimension - 0.6
Receptor range - 180
Ligand range - 180
Twist range - 360
Distance range - 10
Results
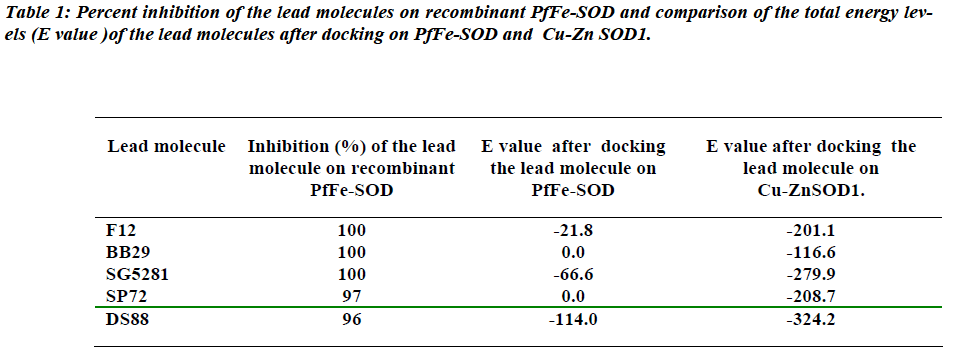
The best fit interactions were found only in five (SP12, BB29, SG5281, DS88, SP72) of the fourteen molecules that were drawn in this study (Fig. 2). The results indi-cate that these ligands do not show much docking on the PfFeSOD but seem to have docking sites or surface inter-actions on the human Cu-ZnSOD1 (Fig. 2).
It appears results of inhibition assays of the lead mole-cules with PfFe-SOD do not show any correlation with respect to docking on the surface of the enzyme. Though F12, BB29, SP72 show no signs of docking on the 3D structure of PfFe-SOD, all the five ligands seem to be interacting with definite pockets on the surface of the 3D structures of the dimeric Cu-ZnSOD1.
Discussion
Our earlier study has shown very little homology between Cu-ZnSOD1 of human and PfFe-SOD, which meant their sequences and structures are very distinct from each other [14]. This would probably be the best basis to target the PfFeSOD for therapy of malaria falciparum caused by this parasite. Due to the phenomenon of drug resistance, a search for new leads against PfFe-SOD has shown that molecules SG5281, F12 and BB29 have 100% inhibition capacity at 50 μM concentration with the recombinant enzyme (7). While, other lead molecules like BB24, BB31, SP112, BB82, DS88, SG7861, SP13 and SP72 show lessor (80-100%) inhibition at that concentration. The best leads among all these were SG5281, F12 and BB29 since they inactivated the recombinant PfFe-SOD enzyme completely at 50 μM concentration. However, these leads behave differently in in vitro assays with two strains of parasite plasmodium falciparum cultures, HB3 (chloroquine sensitive) and Dd2 (moderately chloroquine resistant). Ten of the fourteen were inactive [7]. Of the three leads (SG5281, F12 and BB29) which inactivated the recombinant enzyme completely, two (BB29, F12) seem to be totally inactive, while, SG5281 value showed only 38% inhibition on Dd2 strain. The authors speculate that this difference in the behaviour of the lead molecules between recombinant enzyme and parasite cultures, maybe due to either decreased stability of molecules dur-ing assay or degradation of the molecules before they reach the target or absence of in vivo physiological condi-tions in the assay. Another reason for the altered behavior in cultures could be because the lead molecules may not have bound to the native enzyme. This difference in the behaviour of lead molecules with the recombinant en-zyme and in vitro culture assays clearly indicates a need for lead molecules to be validated by alternate methods before they could be made / called as drug leads. More-over, the IC50 values of these leads are in micromolar range as compared to the current drug of choice chloro-quine which is in nanomolar range (21.2 nM and 54.9 nM for HB3 and Dd2 respectively) which is a thousand fold higher [7]. The study however, does not in any way indi-cate the effect of these lead molecules on the host Cu-ZnSOD1. In vitro inhibition assays of the lead molecules with the recombinant Cu-ZnSOD1 could also be done to check out possible interaction leading to inhibition. Al-though the results may be inconclusive as in the case of the Hb3 and Dd2 strain cultures of the parasite, it would probably throw some light on their effect on the enzyme.
A recent but alternate method to check out possible inter-action between lead molecules and target enzyme is in-silico docking. In recent times, the insilico approach to drug designing is gaining potential significance in en-hancing the process of drug discovery [13]. This approach is an attempt to streamline the lead discovery process by moving as much of the in vitro experimentation as possi-ble into the realm of computation, with the help of the ligand (lead molecule) docking and screening algorithms. Insilico docking of leads with 3D structures of target en-zymes will throw light on whether the target enzyme has docking sites / pockets for the lead. This could be the possible alternate method for validation of drug leads in conjunction with the in vitro assays with recombinant enzymes and cultures of the parasite.
The results of the docking studies indicate that there are no docking sites on PfFe-SOD for these lead molecules which showed considerable inhibition with the recombi-nant enzyme. This could probably be the reason why these molecules did not show inhibition with the plasmo-dium falciparum cultures [14]. The E-values from the docking studies suggest that, the ligand–Cu-ZnSOD1 in-teraction would probably be more favorable than ligand-PfFe-SOD interaction. Even though the in vitro SOD in-hibition assay of the ligands with recombinant PfFe-SOD showed inhibition (96-100%) of the enzyme, the insilico docking studies and the lower E values clearly show a better interaction with the host Cu-ZnSOD1. Taking this into consideration, it would be advisable to check the IC50 value of lead molecules with recombinant Cu-ZnSOD1 as well. In addition, an insilico docking evaluation of the possible ability of the lead molecules to dock on the sur-face of the host / parasite SOD could also be used to screen the molecules. An unnecessary interaction of the lead molecule with Cu-ZnSOD1 would decrease its activ-ity in vivo and this could lead to oxidative stress. In con-clusion, this study shows that compounds which are found to be quite effective inhibitors in vitro (such as in assays with recombinant target enzymes / parasite cultures), need to be validated by alternate methods. Often it so happens that in the living system, compounds tend to deviate from their in vitro behavior in an unpredicted way. Thus com-putational tools such as insilico docking provide the sci-entist with an alternate base for validation of lead mole-cules since most of them could cause oxidative stress in the host which could be lethal.
Abbrevations
Superoxide dismutase -SOD; Plasmodium falcifarum iron superoxide dismutase - PfFe-SOD; human cytosolic cop-per-zinc superoxide dismutase - Cu-ZnSOD1; Mitochon-drial manganese superoxide dismutase - Mn-SOD2
References
- Schwartz E, Samuni A, Friedman I, Hempelmann E, Golenser J. The role of superoxide dismutation in ma-laria parasites. Inflammation. 1999; 23(4): 361-70.
- Weisiger RA, Fridovich I. Superoxide Dismutase- Organelle specificity. The journal of Biological chem-istry. 1973; 248 (10): 3582-3592.
- Bozdech Z, Ginsburg G. Antioxidant defense in Plas-modium falciparum-data mining of the transcriptome. Malaria Journal. 2004, 3:1475-2875.
- Sylke Muller. Redox and antioxidant systems of the malaria parasite Plasmodium falciparum. Molecular Microbiology. 2004; 53 (5):1291-1305.
- Parker MW, Blake CC. Iron- and manganese-contain-ing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 1988; 229(2):377-82.
- McCord JM, Fridovich I. Superoxide dismutase: the first twenty years (1968-1988). Free Radic. Biol. Med. 1988; 5(5-6): 363-369.
- Laurent Soulere, Patrick Deplace, Elisabeth Davioud-Charvet, Sandrine Py, Christian Sergheraert, Jacques Perie, Isabelle Ricard, Pascal Hoffmann and Daniel Dive. Screening of Plasmodium falciparum Iron Su-peroxide Dismutase inhibitors and accuracy. Bioor-ganic & Medicinal Chemistry. 2003;11(23): 4941-4944.
- “PubMed: Online search engine for science and bio-medical articles, www.pubmedcentral.nih.gov
- “The Protein Data Bank”. Nucleic Acids Research. 2000; 28(1): 235-242.
- David.W.Rithcie . Evaluation of Protein Docking Pre-dictions using Hex 3.2 in CAPRI rounds 1-2. Proteins, Structure, Function and Genetics. 2003; 52(1): 98-106.
- D.W. Rithcie and G.J.L.Kemp. Protein Docking Using Spherical Polar Fourier Correlation. PROTEINS: Struct. Funct. Genet. 2000; 39(2): 178-194.
- Mathew AJ, Nixon RN. Docking Studies on Anticancer Drugs For Breast Cancer using Hex. Proceedings of the International MultiConference of Engineers and Com-puter Scientists 2009 Vol I; IMECS 2009, March 18 - 20, 2009, Hong Kong.
- Joseph PS, John CW. Docking study of triphenylphos-phonium cations as estrogen receptor and modulator. Bioinformation; 2009 ; 3(7):303-307
- Bernhardt VG., Pinto JRT, Pai VR.. Superoxide Dis-mutase: An alternate target for Plasmodium. Biomedi-cal Research. 2009; 20(2): 127-135.