ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 21
Immunodetection and risk assessment for Aspergillus contamination in nuts using a highly specific monoclonal antibody
Alaa A. Al-Rifaie, Marwan Y. Al-Maqtoofi*
Department of Biology, College of Science, University of Basrah, Basrah, Iraq
- *Corresponding Author:
- Marwan Y. Al-Maqtoofi
Department of Biology
College of Science
University of Basrah, Iraq
Accepted date: November 26, 2018
DOI: 10.4066/biomedicalresearch.29-18-1098
Visit for more related articles at Biomedical ResearchFungal contamination in nuts is unavoidable and is a major challenge to nuts safety, quality and then for human health after consumption particularly immunocompromised individuals. This study aimed to early detection of potential pathogenic Aspergillus in nuts to avoid the harmful effects of allergens and mycotoxins. Three different geographic locations in Basra, Iraq were screened, and a total of 28 fungal isolates were recovered from tested nuts including cashews, almonds, walnuts, pistachio and sunflower seeds. ELISA was used to identify isolated fungi based on antigenic structure via using a highly specific MAb for Aspergillus, JF5. The accuracy of ELISA was validated by ITS-rDNA sequence in comparison with NCBI database. The highest incidence of isolated fungi belongs to main four genera Aspergillus (78.57%), Alternaria (10.71%), Cladosporium (7.14%), followed by Penicillium (3.75%). The most dominant species was A. flavus, a potential aflatoxins producer, (28.57%). Abu was highly contaminated geographic site comparing with Zubayr and Qarmat. These findings raise awareness about contaminated nuts particularly there is no database available and highlighted the accuracy of using specific MAb for fungal identification and diagnosis. The occurrence of well-known toxigenic species such as A. niger, A. flavus and A. terreus suggested the possible risk of mycotoxin contamination of the nuts for human consumption.
Keywords
Monoclonal antibody, Fungal antigen, Aspergillus, ITS, Diagnosis, Nuts
Introduction
Nuts are widely consumed as plant origin foods due to the nutrition they contain, and are considered a source of microorganism growth, including toxigenic and pathogenic fungal species. Therefore, contamination of nuts has a significant impact in plant production, economics, human and animal health [1]. The expanded nut-products trade has a highlighted nut’s role in the exposure to diseases associated with the occurrence of toxigenic fungal species and their secondary metabolites [2].
Mycotoxigenic fungi are ubiquitous environmental moulds, and pathogens of both plants and animals [3]. These pathogens are considered as natural contaminants of nuts. The most predominant mycotoxic and pathogenic fungi are in tropical environments, and mainly belong to Fusarium, Aspergillus, Penicillium, and Alternaria [4]. Moreover, these fungi have been reported as serious life-threatening pathogens with high rates of mortality and morbidity in immunocompromised patients [5,6]. Nut contamination can occur either by fungal colonization or their production of secondary metabolites during or after harvesting, storage and transition [7].
Mycotoxins are fungal secondary metabolites and exhibit toxic effects in human and animal health through contaminant foods or feeds consumption [8]. The presence of mycotoxins in nuts leads to a disease called mycotoxicosis that has emerged in several countries and requires development of new strategies to early prevention, also require urgent investigations to avoid human infections [9]. Since Aspergillus species, including A. flavus, A. niger, A. fumigatus, A. terreus and A. nidulans, are a major wide spread source of aflatoxins (AFTs) production [10], these pathogenic fungi are one of the biggest threat to human health. The risky aspects of these species are due to invasive aspergillosis, leading to high mortality rate in impaired immune response individuals via direct propagules infection [11,12], or in aflatoxicosis through of aflatoxin-tainted foods [13]. Liver cancer, reduction in protein metabolism, mutagenic, skin necrosis and fat absorption are all risky consequences of contaminated nuts [14]. For the immune system, aflatoxins significantly supress T-cell activities and macrophage phagocytosis, resulting in immunosuppression [15-17].
There is no formal information about nuts consumption in the Iraqi population comparing with other countries available by the Food and Agriculture Organization (FAO) or any of academic study. We understand the consumption of nuts in Iraq is a kind of daily habits. Based on this fact, the rate of nuts consumption is estimated to be ranged between 50 to 150 g/ person daily. Sunflower seeds the most popular kind of nuts consume daily. Generally, there is no data available about the role of contaminated nuts and infected patients after nuts consumption. Recent studies have shown an urgent need for mycotoxigenic fungi identification globally to develop a mycotoxins prevention strategy [18].
To our knowledge, there is no study has used the combination of highly specific monoclonal antibody (MAb) JF5 and molecular technique Internal Transcribed Spacer (ITS) for diagnosis Aspergillus species isolated from contaminated nuts for human consumption. This work aimed to screen nuts and determine the potential fungal contamination using immunodiagnostic test to provide an epidemiology view of opportunistic pathogens that can be associated with mycoses infections due opportunistic pathogens. The study is focused on three geographical areas that close to borders and have the largest portion of population in Basrah, South of Iraq.
Material and Methods
Sample collection
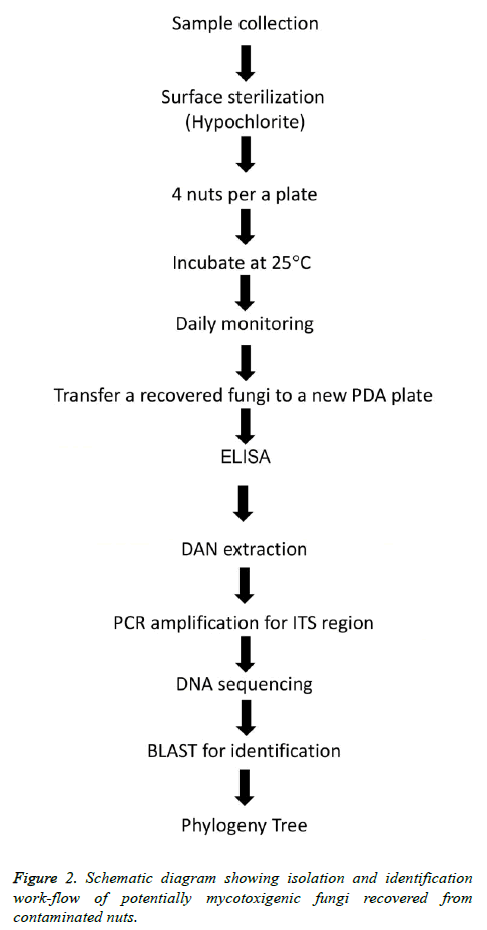
Five kinds of unroasted and unsalted nuts including cashew, almonds, walnuts, pistachio and sunflower seeds were purchased by laboratory staff (1 kg) each type between April to May 2017 from three different geographic local markets in Abu Al Kasib, Qarmat Ali and Zubayr within Basrah province, Iraq (Figure 1). The first three types of nuts were unshelled whereas pistachio nuts were split-shells and sunflower seeds were in-shell. Nuts samples were immediately transferred to the laboratory as soon as purchased, for processing on arrival by mycological culture and molecular treating as illustrated in (Figure 2).
Fungal isolation from nuts
To study the mycotoxigenic fungi of the nuts samples, 90 seeds of each kind of nuts were placed on petri dishes and then superficially sterilized by flooding with 5% sodium hypochlorite solution freshly prepared for 5 min at room temperature followed by washing three times with sterile H2O. Washed samples were then blotted on sterile Whatman filter paper sheets to dry prior cultivation. Four seeds of each nuts kind at three replicates were then placed on the surface of potato dextrose agar (PDA) (LAB098; Lab M Limited, UK) containing 0.1 mg/ml of the broad-spectrum antibiotic Rifampicin (P17278; Ajanta Pharma, India). The plates were then incubated at 25°C and daily monitored for fungal growth recovery. A sample was considered infected when fungal growth was detected. Subsequently, the recovered isolates were purified by sub-culturing on PDA with rifampicin until axenic cultures were generated.
A reference code was allocated to each of the 28 isolates recovered. The isolates were then maintained as PDA slopes at 25°C before DNA extraction for molecular identification of fungi as described below.
For Aspergillus antigen detection, recovered fungi were grown on PDA slopes and surface washing containing water-soluble antigens prepared as described in [19]. Fifty μl volumes were then used to coat the wells of microtitre plates. After incubating overnight at 4°C, wells were washed four times with PBST (PBS containing tween-20, 0.05% (v/v)), once each with PBS and distilled H2O and then air-dried in a laminar flow hood. Wells containing immobilized antigens were blocked for 10 min with 100 μl of blocking solution 1% (wt/ vol) BSA (Sigma A-7030) in PBS. Diluted primary MAb JF5 (mouse IgG3 specific to Aspergillus and Penicillium spp.) [20] in 10% FBS/PBS (1:1000) was conjugated with HRP, following manufacture instructions from Lightning-LinkTM HRP Conjugation Kit (Expedeon, UK, #701-0001), and then was added into the wells for 1 h incubation. Bound antibody was visualised by incubating wells with TMB (T2885; Sigma) substrate solution for 30 min. The reactions were stopped by the addition of 3 M H2SO4 and absorbance values were determined at 450 nm. Wells were given four 5 min rinses with PBST between incubations and a final rinse with PBS before addition of the substrate solution. Working volumes were 50 μl per well and control wells were incubated with PBS. All incubation steps were performed at 25°C in sealed plastic bags. The threshold for detection of the antigen in ELISA was determined from control means. Consequently, absorbance values>0.100 were considered as positive for the detection of antigen.
Identification of fungi by analysis of the ITS regions of the rRNA-encoding gene unit
The recovered fungi were grown on PDA for 5 d at 25°C and then 3 × 5 mm agar plugs taken from the leading edge of PDA cultures, inoculated into 250 ml conical flasks containing 100 ml of potato dextrose broth (PDB), and incubated at 25°C for 48 h. Fungal biomass were collected by filtration through a sterile Whatman filter paper and snapped in liquid N2 (LN2). Collected mycelia were ground in a mortar with LN2 until a fine powder was produced and then transferred into a clean 1.5 ml microcentrifuge. Genomic DNA extraction was conducted using hexadecyltrimethylammonium bromide (CTAB) method as described in [21]. Briefly, 500 μl of 2X hexadecyltrimethylammonium bromide (CTAB) was added, incubated for 30 min at 65°C with occasional shaking. An equal volume of chloroform:isoamyl:alcohol (24:1:1) was added and shaken for 30 min at room temperature. The samples were then centrifuged at 14000 Xg for 10 min.
The supernatant was collected into clean tubes followed by adding 500 μl of chilled isopropanol and incubation on ice for 5 min. Next, samples were centrifuged at 14000 Xg for 10 min and the pellet was drained and resuspended in 500 μl in nuclease-free-water. The DNA was re-precipitated by adding 0.1 vol of sodium acetate, 3 M (pH 5.2) and 2 vol of absolute ethanol and incubated for 10 min at 20°C. After centrifugation at 14000 Xg for 20 min, the purified genomic DNA was recovered and washed with 400 μl of ethanol, 70% (v/v), and then was dried for 5 min in a vacuum rotary desiccator and resuspended in 25 μl of nuclease-free water (Sigma) containing RNAse A, 10 μg/ml. A NanoDrop spectrophotometer (Thermo Scientific) was used to quantify DNA concentration.
PCR and gel electrophoresis
Sequencing of the ITS1-5.8S-ITS2 region of the rRNAencoding gene unit technique was used for recovered mycotoxigenic fungi identification at molecular level according to procedures described elsewhere [5], using the primers ITS1ext (5’-GTAACAAGGTTTCCGTAGGTG-3’) and ITS4 ext (5’-TTCTTTTCCTCCGCTTATTGATATGC-3’). The primer sequences are based on those described by White et al. [22]. The ITS1-5.8S-ITS2 region was amplified by polymerase chain reaction (PCR) procedure using PT-100 thermocycler as follows. The PCR mixture was carried out using a gDNA template approximately 10-25 ng, 25 μl Red Taq Master Mix (VWR), 2.5 μl of each primer at 0.5 picomoles (pmol), and the final volume of reaction was adjusted to 50 μl with Nuclease free water or MQ water. The 37 cycles of PCR conditions after a first initial denaturation step at 95°C for 5 min were followed: 94°C for 30 s, 55°C for 30 s, 72°C for 1.5 min, and final extension at 72°C for 10 min.
To confirm amplification and the gDNA band size, the PCR products were subjected to electrophoresis in 1.5% agarose gel (ApplyChem) using a 1X tris-borate EDTA buffer (TBE) (0.09 M tris-borate, 2 mM EDTA) with the addition of ethidium bromide 4 μl of a 0.5 μg/ml stock per 100 ml of agarose. The amplified DNA band size was determined using the 1 Kb plus size marker (Invitrogen). DNA bands were visualized and imaged under UV light using UV lamp (Spectroline). After DNA fragments were cut from gel, amplified bands were purified using PCR DNA clean kit (Thermo Fisher) according to the manufacturer’s instructions.
Nucleotide sequence accession numbers
Newly determined ITS sequences were submitted to GenBank and the ITS accession numbers MH237623 to MH237650 were obtained. Species designations of recovered fungi are shown in Table 1.
| Location | Isolate code | Source | Predicted fungal genus | ELISA Absorbance | Identification based on | GeneBank |
|---|---|---|---|---|---|---|
| Ab at 450nm | ITS sequencing | Accession no. | ||||
| Abu Al Kasib | A2 | Walnut | Aspergillus | 0.943 | Aspergillus sp. Mar1 | MH237623 |
| Abu Al Kasib | A3 | Walnut | Aspergillus | 1.211 | Aspergillus flavus | MH237624 |
| Abu Al Kasib | A4 | Sunflower Seeds | Aspergillus | 0.974 | Aspergillus oryzae | MH237625 |
| Abu Al Kasib | A5 | Sunflower Seeds | Aspergillus | 1.065 | Aspergillus nidulans | MH237626 |
| Abu Al Kasib | A6 | Sunflower Seeds | Alternaria | 0.001 | Alternaria alternata | MH237627 |
| Abu Al Kasib | A7 | Cashew | Penicillium | 0.627 | Penicillium griseofulvum | MH237628 |
| Abu Al Kasib | A8 | Cashew | Aspergillus | 1.203 | Aspergillus flavus | MH237629 |
| Abu Al Kasib | A9 | Cashew | Cladosporium | 0.002 | Cladosporium herbarum | MH237630 |
| Abu Al Kasib | A10 | Cashew | Aspergillus | 1.192 | Aspergillus flavus | MH237631 |
| Abu Al Kasib | A11 | Pistachio | Aspergillus | 1.253 | Aspergillus flavus | MH237632 |
| Abu Al Kasib | A17 | Amond | Aspergillus | 1.099 | Aspergillus flavus | MH237633 |
| Abu Al Kasib | A18 | Cashew | Aspergillus | 1.381 | Aspergillus niger | MH237634 |
| Qarmat Ali | K3 | Walnut | Aspergillus | 1.27 | Aspergillus niger | MH237635 |
| Qarmat Ali | K5 | Cashew | Aspergillus | 1.001 | Aspergillus sp. Mar2 | MH237636 |
| Qarmat Ali | K6 | Cashew | Aspergillus | 1.089 | Aspergillus flavus | MH237637 |
| Qarmat Ali | K7 | Pistachio | Aspergillus | 1.209 | Aspergillus niger | MH237638 |
| Qarmat Ali | K8 | Sunflower Seeds | Aspergillus | 0.554 | Aspergillus ruber | MH237639 |
| Zubayr | Z1 | Cashew | Aspergillus | 1.019 | Aspergillus flavus | MH237640 |
| Zubayr | Z2 | Cashew | Aspergillus | 1.256 | Aspergillus niger | MH237641 |
| Zubayr | Z3 | Cashew | Aspergillus | 1.046 | Aspergillus terreus | MH237642 |
| Zubayr | Z4 | Cashew | Alternaria | 0.002 | Alternaria alternata | MH237643 |
| Zubayr | Z5 | Cashew | Alternaria | 0.001 | Alternaria infectoria | MH237644 |
| Zubayr | Z6 | Sunflower Seeds | Cladosporium | 0.002 | Cladosporium limoniforme | MH237645 |
| Zubayr | Z8 | Almond | Aspergillus | 1.28 | Aspergillus terreus | MH237646 |
| Zubayr | Z9 | Almond | Aspergillus | 1.262 | Aspergillus terreus | MH237647 |
| Zubayr | Z11 | Pistachio | Aspergillus | 1.067 | Aspergillus terreus | MH237648 |
| Zubayr | Z14 | Walnuts | Aspergillus | 1.195 | Aspergillus niger | MH237649 |
| Zubayr | Z15 | Walnuts | Aspergillus | 1.037 | Aspergillus flavus | MH237650 |
Table 1. Results of ELISA detection and identification of fungi using ITS sequencing.
Species identification by DNA Sequence analysis and phylogenetic tree
The purified PCR products were submitted to Macrogen Korea for sequencing. Subsequently, the recovered fungal sequences data were compared with database of the GenBank species by depositing in the National Center for Biotechnology Information (NCBI) databases by using NCBI BLAST® (https://blast.ncbi.nlm.nih.gov). According to Leonard et al. [23], a serial of bioinformatics tools were used for basic editing and analysis to establish the phylogenetic tree including SeaView Version 4.5.4 (http://doua.prabi.fr/software/seaview) (Manolo Gouy, Laboratoire de Biométrie et Biologie Evolutive CNRS/ Université de Lyon “Licensed under the GNU General Public Licence”), FigTree Version 1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) (Andrew Rambaut Institute of Evolutionary Biology, University of Edinburgh), REFGENE bioinformatics tool (http://richardslab.exeter.ac.uk/refgen.html) and TREENAMER bioinformatics tool (http://richardslab.exeter.ac.uk/treenamer.html).
Results
Isolation and Frequencies of fungal contaminants
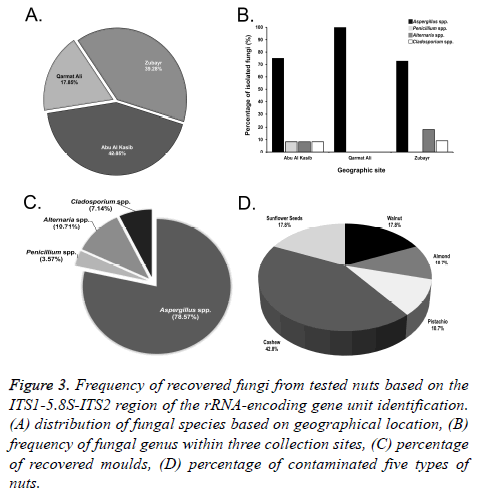
In this study, a total of 60 samples of unroasted and unsalted nuts, in triplicate (20 samples each replicate) were tested. Nuts were examined to detect the percentage of potential mycotoxigenic fungal contamination. Based on the geographical distribution, nut samples from Abu Al Kasib (42.85%) were the most contaminated commodities by fungal species, followed by Zubayr (39.28%) and Garmat Ali (17.85%) (Figures 3A and 3B).
Figure 3: Frequency of recovered fungi from tested nuts based on the ITS1-5.8S-ITS2 region of the rRNA-encoding gene unit identification. (A) distribution of fungal species based on geographical location, (B) frequency of fungal genus within three collection sites, (C) percentage of recovered moulds, (D) percentage of contaminated five types of nuts.
A total of 28 fungal species were recovered from nuts that surface-sterilized. The incidence of Aspergillus species was dominant (71.42%). Both Penicillium spp. and Alternaria spp. were recovered at the same rate (10.71%), compared with the lowest contamination of all commodities by Cladosporium spp. (7.14%) (Figure 3C).
Figure 3D shows the percentage of unroasted and unsalted five nut samples that were naturally contaminated by fungi. Cashews were found to be the most significantly contaminated nuts (42.85%), with potential mycotoxigenic fungal species of Aspergillus, Penicillium, Alternaria and Cladosporium, while the lowest level of contamination was found in both almonds and pistachios (10.71%). Walnuts and sunflower seeds were in the middle (17.85%).
Immunodetection of fungal antigens
ELISA test using highly specific MAb, JF5, to Aspergillus and related fungi belong to certain Penicillium species was used to identify recovered axenic fungal cultures from nuts via tracking crude antigen extracts. The MAb was shown to be specific for their target species (Table 1). Total of 28 fungal isolates tested by ELISA, up to 71% reacted with MAb JF5 specific for Aspergillus. The remaining fungi were unrelated species non-reactive with MAb JF5. In addition, MAb crossreact with related with antigens from Penicillium griseofulvum.
Identification of fungal species
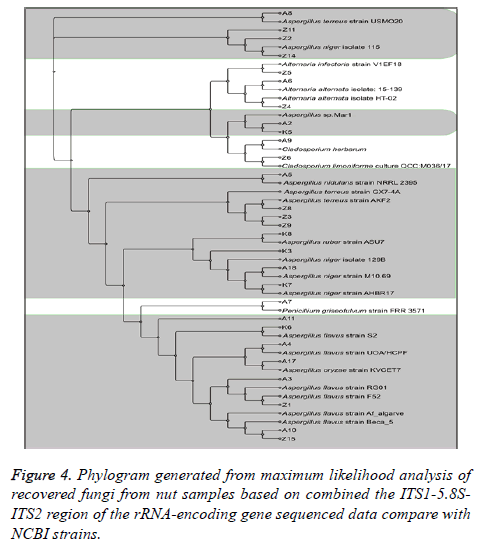
The result of DNA sequencing analysis of ITS region for 28 fungal isolates confirmed the specificity test of ELISA outcomes. MAb JF5 successfully detect strains of A. flavus prevalence was (28.57%), followed by A. niger (17.85%). A. nidulans and A. ruber were less frequent at (3.57%). An undefined Aspergillus was isolated (10.71%) at species level after NCBI submission. Other fungal species recovered from tested commodities include: Alternaria alternata, Alternaria infectoria, Cladosporium herbarum, Cladosporium limoniforme and Penicillium griseofulvum (Table 1 and Figure 4).
Discussion
Mycotoxins have great impact on the human health, animal production and countries economy [24]. Similar to the other crop plants, nuts are susceptible to the growth of mycotoxin producing fungi and contain significant amount of different mycotoxins, most prominently, aflatoxins produced by Aspergillus species [25]. These toxigenic fungi are ubiquitous and have a particular importance as nuts contaminants of human consumption. In addition to the reduction of nutrient in contaminated nuts, the activity of fungal propagules growth and mycotoxin production indicates a potential increase risk in developing pulmonary mycotoxicosis and aspergillosis, particularly in immunocompromised patients [26]. The most common Aspergillus species associated with aflatoxins production is A. flavus [27]. This species is widely detected in food and feeds stuffs [28].
Additionally, aflatoxins have reported as a group 1 human carcinogen by the International Agency for Research on Cancer [14]. They also have bioaccumulation property leading to teratogenic [29].
Since aflatoxins are serious immunosuppressive mycotoxins, consumption of contaminated nuts with aflatoxins results in dysregulation of the immune cells: proliferating, differentiating, and the communication network between cellular and humoral components is disrupted. In addition, mycotoxin-induced immunosuppression may lead to impaired T-lymphocytes and B-cells, and specifically to antibody production. Significant effect of these toxins has been reported mainly through the suppression of phagocytosis activity of innate immune cells, including macrophages and neutrophils [30-32]. Due to the defects in human immune responses after consuming mycotoxin contaminated commodities, the susceptibility to opportunistic fungal infections such aspergillosis is increased [33].
Extensive research on mycotoxins contamination production, detection, distribution, and their medical effects in agriculture products, including nuts, have been done worldwide such the US, Brazil, India, Nigeria, and others. The main goal of this study was to evaluate the mycotoxigenic contamination in nuts for human consumption, and then better understand their distribution. To the best of our knowledge, this study has demonstrated, for the first time, the detection and distribution of toxigenic fungi using this molecular technique, ITS-rDNA, in nuts intended for human consumption within three different local geographic areas in the south part of Iraq, Basra.
In this study, a total of 28 different potential mycotoxigenic fungi were recovered from nut samples, which include shelled cashews, almonds, walnuts, half-shelled pistachios, and unshelled sunflower seeds. These commodities were selected in this study as they are the most popular nut products consume by both competent and immune-impaired individuals. All isolated species were filamentous fungi with no observation to yeast moulds. These isolates were potential toxigenic species belong to main four genera that well known as mycotoxins producers and human pathogens including Aspergillus, Penicillium, Alternaria and Cladosporium. Previous studies have reported similar observation of fungal contamination in tested commodities [34,35]. On the other hand, yeasts have been isolated from contaminated nuts at a very low percentage [36,37].
The results of this study show that the highest rate of nut contamination came from Aspergillus species (78.57%) followed by Alternaria, Cladosporium and Penicillium.
Significantly, A. flavus was the most prevalent contaminant species in all tested nut samples including cashews, walnuts, sunflower seeds, almond and pistachios in accordance with [38]. This species is ubiquitous and has capability to produce one or more of the carcinogenic aflatoxins that causing aflatoxicosis after human consumption, in particular by immunocompromised patients [27,39]. The International Agency for Research on Cancer has identified aflatoxin as group 1, highly carcinogenic product, to human [14].
The percentage of nut contamination by fungal propagules varies based on nut type. The analysis test of fungal recovery showed that cashews were the most loaded nuts with fungal contamination (42.85%), compared with the other samples. Importantly, the incidence of A. flavus was the most commonly identified species in cashews [40], this finding is similar to our observation. Shells of nuts are the first physical barrier that preventing microbial invasion that explains the susceptibility of unshelled nut samples to fungal contamination [41]. In addition to the matrix structure of cashew, the moisture and nutrient contains including carbohydrates, proteins and lipids that encourage fungal propagules growth, penetration and mycotoxin production [42].
Based on geographic location, purchased nuts from Abu Al Kasib were the most contaminated samples followed by Zubayr and lastly Qarmat Ali. The 100% accuracy of the immunodiagnosis test using MAb JF5 ELISA, confirmed by using ITS sequencing PCR analysis of recovered isolates, demonstrates its robustness in detecting potentially toxigenic Aspergillus species in contaminated nuts. Around two third of axenic isolates were Aspergillus species [20]. Many factors have influenced toxigenic fungi for mycotoxin products, for example, storage conditions and ways of transport. High level of humidity during storage can stimulate mycotoxin production [2].
Acknowledgement
Authors thank all participants for their cooperation in this study.
References
- Cleveland TE, Dowd PF, Desjardins AE, Bhatnagar D, Cotty PJ. United States Department of Agriculture-Agricultural Research Service research on pre-harvest prevention of mycotoxins and mycotoxigenic fungi in US crops. Pest Manag Sci 2003; 59: 629-642.
- Moretti A, Logrieco AF, Susca A. Mycotoxins: an underhand food problem. Methods Mol Biol 2017; 1542: 3-12.
- Thornton CR, Wills OE. Immunodetection of fungal and oomycete pathogens: established and emerging threats to human health, animal welfare and global food security. Crit Rev Microbiol 2015; 41: 1-25.
- Baquião AC, De Oliveira MMM, Reis TA, Zorzete P, Atayde DD, Correa B. Monitoring and determination of fungi and mycotoxins in stored Brazil nuts. J Food Protect 2013; 76: 1414-1420.
- Al-Maqtoofi M, Thornton CR. Detection of human pathogenic Fusarium species in hospital and communal sink biofilms by using a highly specific monoclonal antibody. Environ Microbiol 2016; 18: 3620-3634.
- Bilgrami KS, Choudhary AK. Competing mycoflora with Aspergillus flavus in kernels of Rabi and Kharif maize crops of Bhagalpur. Ind Phytopathol 1990; 43: 547-452.
- Wild CP, Gong YY. Mycotoxins and human disease: a largely ignored global health issue. Carcinogenesis 2010; 31: 71-82.
- Bacha H, Hadidane R, Creppy EE, Regnault C, Ellouze F, Dirheimer G. Monitoring and identification of fungal toxins in food products, animal feed and cereals in Tunisia. J Stored Prod Res 1988; 24: 199-206.
- Paterson R, Lima N. Filamentous fungal human pathogens from food emphasising Aspergillus, Fusarium and Mucor. Microorganisms 2017; 5: 44.
- Chakrabarti A, Chatterjee SS, Das A, Shivaprakash MR. Invasive aspergillosis in developing countries. Med Mycol 2011; 49: 35-47.
- Shephard GS. Impact of mycotoxins on human health in developing countries. Food Additives Contaminants Part A 2008; 25: 146-151.
- Ostry V, Malir F, Toman J, Grosse Y. Mycotoxins as human carcinogens-the IARC Monographs classification. Mycotoxin Res 2016; 33: 65-73.
- Bennett JW, Klich M. Mycotoxins. Clin Microbiol Rev 2003; 16: 497-516.
- Fokunang CN, Tembe-Fokunang EA, Tomkins P, Barkwan S. Global impact of mycotoxins on human and animal health management. Outlook Agric 2006; 35: 247-253.
- Pitt JI. Toxigenic fungi and mycotoxins. Br Med Bull 2000; 56: 184-192.
- Dawlatana M, Shahida S, Rahim M, Hassan MT. Investigation on the occurrence of ochratoxin A in Maize in Bangladesh. Bangl J Sci Industr Res 2008; 43: 495-500.
- Thornton CR. Immunological methods for fungi. Molecular and cellular biology of filamentous fungi: a practical approach. Oxford Univ Press 2001; 227-257.
- Thornton CR. Development of an immunochromatographic lateral-flow device for rapid serodiagnosis of invasive aspergillosis. Clin Vaccine Immunol 2008; 15: 1095-1105.
- Chow TYK, Kafer E. A rapid method for isolation of total nucleic acids from Aspergillus nidulans. Fungal Genet Newsl 1993; 40: 25-27.
- White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. USA: Academic Press 1990; 315-322.
- Leonard G, Stevens JR, Richards TA. REFGEN and TREENAMER: automated sequence data handling for phylogenetic analysis in the genomic era.
- Evol Bioinform Online 2009; 5: 1-4.
- Darwish WS, Ikenaka Y, Nakayama SM, Ishizuka M. An overview on mycotoxin contamination of foods in Africa. J Vet Med Sci 2014; 76: 789-797.
- Kishore GK, Pande S, Manjula K, Rao JN, Thomas D. Occurrence of mycotoxins and toxigenic fungi in groundnut (Arachis hypogaea L.) seeds in Andhra Pradesh, India. Plant Pathol J 2002; 18: 204-209.
- Pagano L, Akova M, Dimopoulos G, Herbrecht R, Drgona L, Blijlevens N. Risk assessment and prognostic factors for mould-related diseases in immunoco-mpromised patients. J Antimicrob Chemother 2011; 66: 5-14.
- Jeswal P, Kumar D. Mycobiota and natural incidence of aflatoxins, ochratoxin A, and citrinin in Indian spices confirmed by LC-MS/MS. Int J Microbiol 2015.
- Streit E, Naehrer K, Rodrigues I, Schatzmayr G. Mycotoxin occurrence in feed and feed raw materials worldwide: long-term analysis with special focus on Europe and Asia-Streit. J Sci Food Agric 2013; 93: 2892-2899.
- Maresca M, Fantini J. Some food-associated mycotoxins as potential risk factors in humans predisposed to chronic intestinal inflammatory diseases. Toxicon 2010; 56: 282-294.
- Gong Y, Hounsa A, Egal S, Turner PC, Sutcliffe AE, Hall AJ. Postweaning exposure to aflatoxin results in impaired child growth: a longitudinal study in Benin, West Africa. Environ Health Perspect 2004; 112: 1334-1338.
- Khlangwiset P, Shephard GS, Wu F. Aflatoxins and growth impairment: a review. Crit Rev Toxicol 2011; 41: 740-755.
- Oswald IP, Marin DE, Bouhet S, Pinton P, Taranu I, Accensi F. Immunotoxicological risk of mycotoxins for domestic animals. Food Additives Contaminants 2005; 22: 354-360.
- Pierron A, Alassane-Kpembi I, Oswald IP. Impact of mycotoxin on immune response and consequences for pig health. Anim Nutr 2016; 2: 63-68.
- Adebajo LO, Diyaolu SA. Mycology and spoilage of retail cashew nuts. African J Biotechnol 2003; 2: 369-373.
- Jogee SP, Ingle AP, Gupta IR, Bonde SR, Rai MK. Detection and management of mycotoxigenic fungi in nuts and dry fruits. Acta Horticulturae 2012; 69-77.
- Nidaa SH, Abeer FM, Eman AA. Molecularly diagnostic of aflatoxigenic Aspergillus flavus isolated from nuts. Res J Env Toxicol 2016; 10: 39-49.
- Tournas V, Niazi N, Kohn J. Fungal presence in selected tree nuts and dried fruits. Microbiol Insights 2015; 8: 1-6.
- Fakruddin M, Chowdhury A, Hossain MN, Ahmed MM. Characterization of aflatoxin producing Aspergillus flavus from food and feed samples. Springer Plus 2015; 4: 159.
- Peraica M. Toxic effects of mycotoxins in humans. Bull WHO 1999; 77: 754-766.
- Kumar P, Mahato DK, Kamle M, Mohanta TK, Kang SG. Aflatoxins: a global concern for food safety, human health and their management. Frontiers Microbiol 2017; 7: 2170.
- Adeniyi OD, Adedeji AR. Evaluation of fungal flora and mycotoxin potential associated with postharvest handlings of Cashew Nut. Arch Appl Sci Res 2015; 7: 30-33.
- Muni CR, Freirea FC, Soare AA, Cooke PH, Guedes MF. The ultrastructure of shelled and unshelled cashew nuts. Micron 2013; 54-55: 52-56.
- Lamboni Y, Nielsen KF, Linnemann AR, Gezgin Y, Hell K, Nout MJ, Smid EJ, Tamo M, Boekel MA, Hoof JB, Frisvad JC. Diversity in secondary metabolites including mycotoxins from strains of Aspergillus section Nigri isolated from raw cashew nuts from Benin, West Africa. Plos One 2016; 11: 0164310.