ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 15
Genotyping and clinical characteristics screening of children with intellectual disability in Western India
Yashvant Khimsuriya1, Nikhil Kharod2 and Jenabhai Chauhan1*
1Department of Genetics, Ashok & Rita Patel Institute of Integrated Study & Research in Biotechnology and Allied Sciences (ARIBAS), New Vallabh Vidyanagar, Anand, Gujarat, India
2Pediatrics Department, H.M. Patel Center for Medical Care and Education, Karamsad, Gujarat, India
- *Corresponding Author:
- Jenabhai Chauhan
Department of Genetics
Ashok & Rita Patel Institute of Integrated Study & Research in
Biotechnology and Allied Sciences (ARIBAS)
New Vallabh Vidyanagar, Anand, Gujarat, India
Accepted on August 18, 2018
DOI: 10.4066/biomedicalresearch.29-18-700
Visit for more related articles at Biomedical ResearchBackground: The RhoGTPases (ARHGEF6, OPHN1 and PAK3) are key signaling proteins, and can inter-relate extracellular and intracellular signals to sort out changes in the actin cytoskeleton of neuronal network connectivity. IL1RAPL2 is a member of Interleukin 1 receptor family that is expressed at high level in post-natal brain structures involved in the hippocampal memory system. The mutations of these genes have been reported to cause intellectual disability (ID). Therefore, this study is conducted to scrutinize distinctive distributions of genomic variations on West Indian population.
Subjects and Methods: The genotyping for determination of SNPs associated with X-linked genes (rs35747426 in ARHGEF6, rs121434612 in PAK3, rs199985543 in OPHN1 and rs9887672 in IL1RAPL2) was performed in phenotypically screened 116 intellectually disabled and 100 healthy children by PCRRFLP method.
Results: It has identified that allelic frequencies for rs199985543 and rs9887672 are seen progressively present in the disease group, indicating a significant level of association with ID in Gujarat population. The multifactor dimensionality reduction (MDR), SNP-SNP genotype models are resulted with a relationship of ID if there should be an occurrence of the OPHN1 and IL1RAPL2 gene variants. Additionally, rs199985543 was analyzed for OPHN1 protein structure and stability prediction, which results damaging effect to the protein. Surprisingly, no variation was found for rs35747426 ARHGEF6 and rs121434612 PAK3 polymorphisms in the present study population.
Conclusion: This phenotype-genotype relationship has a solid and measurable connection according to the carrier genotypes of the variations and different types of ID, that confirm the mutation in OPHN1 and IL1RAPL2 genes permit serious attention for disease risk.
Keywords
Intellectual disability, Genotype-phenotype distribution, Protein phenotype prediction, MDR analysis, OPHN1, IL1RAPL2.
Abbreviations
RhoGTP: Rho Guanosine Triphosphate; ARHGEF6: Activator of Rho Guanine Nucleotide Exchange Factor 6; OPHN1: Oligophrenin 1; PAK3: Protein Activated Kinase 3; IL1RAPL2: Interleukin-1 Receptor Accessory Protein-Like 2; PCR-RFLP: Polymerase Chain Reaction- Restriction Fragments Length Polymorphism; SNP: Single Nucleotide Polymorphism; GWAS: Genome Wide Association Study; NCBI: National Centre for Biotechnology Information; ExAC: Exome Aggregation Consortium; SIFT: Sorting Intolerant from Tolerant; UCSC: University of California, Santa Cruz; NEB: New England Biolab; μL: Microliter; μM: Micromolar; ml: Milliliter; mM: Millimolar; Min: Minute; ng: Nanogram; S: Second.
Introduction
Intellectual Disability (ID) is a prominent characteristic in most of the neurodevelopmental disorders affecting health, teaching and community services in growing nations. A meta-analysis of international studies reported the prevalence of intellectual disability between 1% and 3% of the worldwide population [1]; the world health organization characterized this clinical condition by Intelligence Quotient (IQ) score of less than 70; mild (55-70), moderate (35-54), severe (20-34) and profound (<20) [2]. The environmental factors such as maternal alcohol abuse during pregnancy, infections, birth complications and extreme malnutrition are major causes of ID. However, genetic factors play a significant role in the development of ID. Hence, there is a need to look for other genetic variations, so that we may improve our understanding of intellectual disability and its risk factors.
It can occur in isolation or combination with congenital malformations or other neurological features such as epilepsy, sensory impairment, autism spectrum disorders (ASD), short stature, skeletal abnormality, facial dysmorphism, head anomalies and its severity (mild, moderate, severe and profound) [3-5].
Referable to the structure as well as the functional complexity of the human brain, the actual pathology of intellectual disability is in a manifold. During development and day-to-day functioning throughout life, various proteins need to be functionally active with the right amount, place and time [6]. The alterations in these dynamic protein interactions lead to problems with cellular processes; including neuronal migration, synaptic functions, neurogenesis and regulation of transcription and translation. These proteins are known to have a significant role in the etiology of ID affecting various cellular signaling pathways [7].
Many research studies identified more than 700 genes for Xlinked and autosomal intellectual disability. Males are more affected by ID found in the worldwide because of their inheritance pattern, and thus chromosome X became a primary focus of research on intellectual disability [8].
One of the most emerging cellular signaling cascades is a RHO GTPase pathway, which consists of guanine-nucleotidebinding proteins that act as ‘molecular switches’. The RHO GTPases-a major group of proteins in ID, are known to cause a wide variety of cellular functions, which are crucial for learning and memory. More than 20 GTPases are likely to be found as effectors, activators and regulators of cell division cycle 42 (CDC42), Ras-related C3 botulinum toxin substrate 1 (RAC1) and RhoA for roles in spine formation and synapse plasticity [9,10]. The interleukin 1 receptor accessory proteins are playing an important role in cognitive function and normal physiology of central nervous system (CNS) through interleukin signaling pathways. The high level of its expression involved in the hippocampal memory system suggests a leading role in the physiological process of memory and learning abilities [11].
The present study involved Rho guanine nucleotide exchange factor 6 (ARHGEF6), p21 protein (Cdc42/Rac) activated kinase-3 (PAK3), Oligophrenin 1-Rho GTPase activating protein (OPHN1) and Interleukin-1 receptor accessory proteinlike 2 (IL1RAPL2) to screen pathogenic variants in the western Indian population. It is therefore not unexpected that the pathogenic variations affecting any one of the genes encoding these proteins can have severe consequences for brain development or cognitive functioning to cause intellectual disability.
India is a vast country, with a population of more than 1.3 billion. The multicultural, multi-ethnic, and metalinguistic nature of India makes it a difficult task for drawing generalizations about intellectual disability. It is due to the paucity of available data sources and their accessibility [12].
Materials and Methods
Sampling groups
Study participants residing in Gujarat region are selected. A total of 116 children having ID with unknown cause, were screened based on their distinct phenotypic appearance and Intelligence Quotient (IQ). These samples were collected from the non-government organizations and other private school/ laboratories. 100 normal children, who never have any health complications, selected from private primary schools as a control group. The age of the children varied between 5 to 18 y. The parents signed Informed consent forms and submitted before sample collection. 2 ml of the blood sample collected for genetic analysis from both the groups.
Clinical measurements and features
All the affected children were subjected to demographic, anthropometric and phenotypic investigations as described in our previous article [13]. Various parameters such as gender, age, height, weight, head circumference and Body Mass Index (BMI) were measured. The birth weight and consanguinity recorded from clinical history.
Bioinformatics approach towards SNPs selection
GWAS Central, NCBI dbSNP, ExAC genome browser, and Human Gene Mutation Database (HGMD) were used to retrieve SNPs of selected genes [14-17]. These SNPs subjected to SIFT and Polyphen 2.0 in silico analysis tools to define pathogenicity of the variants [18,19]. Intronic variants analysis was performed by GWAS 3D web server [20].
Genotyping
The genomic DNA extraction was carried out from peripheral blood leukocytes by standard phenol-chloroform procedure [21]. Four SNPs, rs35747426, rs121434612, rs199985543 and rs9887672, were genotyped using PCR-RFLP methods. Primer sequences were designed using NCBI Primer-BLAST and confirmed by in silico PCR analysis on UCSC genome browser followed by a selection of restriction endonucleases using NEBcutter V2.0 for all the selected variants [22,23].
DNA samples were prepared for PCR with EmeraldAmp® GT PCR master mix (Takara Bio Inc. Japan). Primers sequences synthesized from Sigma-Aldrich Chemical Pvt. Ltd. The aliquot of 100 ng DNA samples was amplified using 10 pMol of primers. The PCR temperature conditions varied with annealing temperature (A). The PCR amplification consisted initial denaturation at 95°C for 5 min, 35 cycles of 94°C for 30 s, A°C for 30 s (Table 1) and 72°C for 40 s, followed by final extension was carried out at 72°C for 5 min.
| SNPs | Primer name sequences (5’ to 3’) | Annealing PCR Product (bp) | Temp. (A’) | Size |
|---|---|---|---|---|
| rs35747426 | ARHGEF6-F-ATCGCGGATGTGGCTTAGTC- | ARHGEF6-R-TGATTGCTGCTCTCTGGTGAT- | 286 | 60°C |
| rs121434612 | PAK3-F-CACCCTCTTAGCTACTTCTGC- | PAK3-R-GTGGCTTACAAAACAAAACAAGGAG- | 309 | 60°C |
| rs199985543 | OPHN1-R-TATCACCAGCAGCATAGAACCC- | OPHN1-F-ATCTCCAAACTCAGAAGGATCTCA- | 372 | 55°C |
| rs9887672 | IL1RAPL2-F-ACCTTTCAGTTTTTCAGTC- | IL1RAPL2-R-CAAGAAGCCATTTGTGTT- | 252 | 55°C |
Table 1. Primers details for PCR amplification.
For RFLP analysis, the PCR product of each SNPs was transferred into separate vials and digested using 2-5 units of specific restriction endonucleases (RE) for 2 to 4 h as per the product manual included. Further details of restriction digestion are given in Table 2. The digested PCR products were separated on 3% agarose gel alongside the 50-bp DNA ladder (Genetix Biotech Asia Pvt. Ltd.). In the case of rs35747426, the mutated DNA has a restriction site for the HaeIII enzyme. Therefore, control DNA was used in every cycle of HaeIII restriction digestion analysis.
| SNPs | RE name | Allele | Incubation (°C/h ) | DNA Pattern (bp) | ||
|---|---|---|---|---|---|---|
| Normal | Carrier | Mutant | ||||
| rs35747426 | HaeIII | T/C | 37/2 | 286 | 286, 189 and 97 | 189 and 97 |
| rs121434612 | HinP1I | C/T | 37/4 | 197 and 112 | 309, 197 and 112 | 309 |
| rs199985543 | BsmF1 | C/A | 37/4 | 178, 117 and 77 | 295, 178, 117 and 77 | 295 and 77 |
| rs9887672 | BsmF1 | C/T | 37/4 | 188 and 64 | 252, 188 and 64 | 252 |
| RE: Restriction Endonuclease; Allele-wild/mutant, rs199985543 variation site at 178 bp on 372 bp PCR product. | ||||||
Table 2. Restriction digestion pattern.
In silico analysis: OPHN1 protein structure and stability predictions
In silico analysis was performed to understand the impact of Ala688Ser amino acid substitution at structure level of OPHN1 protein, we predicted 3D model of this protein using I-Tasser server [24]. We were assessed Ramachandran plot using RAMPAGE server and selected one most favourable protein model from native and mutant models, which were generated by I-Tasser server [25]. We considered the models with highest number of residues in favoured region (RFR).
The structural mutation in OPHN1 protein due to amino acid substitution was computed by primary superimposition of wild and mutant forms of OPHN1 protein models, and the positional RMSD value was calculated among equivalent atom using YASARA tool [26].
The stability analysis of the mutant structure compared with wild type using the support vector machine (SVM) methods, Imutant 2.0, Cutoff Scanning Matrix (mCSM), Site Directed Mutator (SDM) and DUET based prediction of stability change [27,28]. The protein FASTA sequence was used in I-mutant 2.0 tool as well as PDB structure of wild type protein of interest, missense variation details, and chain identifier were provided as an input to the server. The result is in the form of Gibbs free energy (ΔΔG) change, and negative values denote the amino acid change as destabilizing to protein structure.
Statistical analysis
The statistical analysis was performed using data analysis tool in Microsoft Excel program. Categorical data were represented in percentage (%). The comparison between genotype and allelic frequency was made using χ2 test and 2-sided Fisher exact test. The relative risk of the gene with SNP was represented by an odd ratio (OR) and 95% Confidence Interval (CI). Epistasis analysis of the gene-gene interaction of selected SNPs was performed by Multifactor Dimensionality Reduction (MDR) using Generalized Multifactor Dimensionality Reduction software (GMDR Beta 0.9). This method gives a number of output parameters, including cross-validation consistency, the testing balanced accuracy, and the sign test, for all the selected attributes. The cross-validation consistency score is a measure of the degree of consistency with which the selected interaction is identified as the best model among all the possibilities. To assess interactions between genes using this method, the GMDR comprehensive search algorithm (exhaustive search algorithm) was applied, which evaluated all the possible combinations of both SNPs with respect to the risk of ID [29].
Results
Clinical features
In the present study, the distribution of disease groups involved 70% males, which are higher in number compared to the female counterparts. We have calculated the percentage distributions of common clinical features, types of ID and abnormal Body Mass Index (BMI) in children with intellectual disability. For clinical interpretation, we also calculated mean, median and Standard Deviation (SD) for age, weight, and height at examination as well as the birth weight of affected children and summarized in Table 3. The calculated frequencies of observed phenotypes of affected children by the gender (male/female), consanguinity and the reported polymorphisms are also included.
| Clinical features | Male | Female | Consanguinity | OPHN1 (rs199985543) | IL1RAPL2 (rs9887672) |
|---|---|---|---|---|---|
| Male/Female | 80/0 | 0/36 | 6/3 | 1/15 | 3/10 |
| Median age (y) | 10 | 13 | 12 | 11.5 | 13 |
| Average age (y) (SD) | 10.88 (3.79) | 12.58 (3.22) | 12.44 (2.5) | 11.75 (3.66) | 11.6 (3.82) |
| Median birth weight (kg) | 2.27 | 2.5 | 2.5 | 2.6 | 2.25 |
| Average birth weight (kg) (SD) | 2.39 (0.74) | 2.5 (0.77) | 2.53 (0.46) | 2.67 (0.77) | 2.5 (0.82) |
| Median height (cm) | 126.8 | 135.25 | 126 | 129 | 126 |
| Average height (cm) (SD) | 127.2 (23.25) | 131.25 (16.84) | 127.11 (19.26) | 128.6 (20.37) | 127.4 (17.55) |
| Median weight (kg) | 25 | 29 | 24.5 | 25 | 25 |
| Average weight (kg) (SD) | 27.67 (12.25) | 28.86 (9.28) | 27.39 (7.9) | 27 (10.14) | 27.5 (11.62) |
| Features | +/- | +/- | +/- | +/- | +/- |
| Microcephaly | 25/55 | 11/25 | 2/7 | 3/13 | 3/10 |
| Macrocephaly | 26846 | 3/33 | 1/8 | 1/15 | 3/10 |
| Seizure | 32/48 | 16/20 | 4/5 | 6/10 | 8/5 |
| Speech abnormality | 40/40 | 18/18 | 3/6 | 9/7 | 5/8 |
| Autistic disorder | 15/65 | 5/31 | 1/8 | 5/11 | 2/11 |
| Short stature | 25/55 | 15/21 | 4/5 | 7/9 | 5/8 |
| Skeletal abnormality | 42/38 | 15/21 | 3/6 | 8/8 | 5/8 |
| Oral cavity defects | 22/58 | 11/25 | 2/7 | 6/10 | 5/8 |
| Facial dysmorphism | 41/39 | 18/18 | 3/6 | 7/9 | 6/7 |
| CNS anomalies | 21/59 | 4/32 | 2/7 | 4/12 | 2/11 |
| Mild | 44/36 | 13/23 | 2/7 | 4/12 | 5/8 |
| Moderate | 28/52 | 19/17 | 5/4 | 10/6 | 6/7 |
| Severe | 8/72 | 4/32 | 2/7 | 1/15 | 2/11 |
| Under weight | 23/57 | 13/23 | 3/6 | 7/9 | 5/8 |
| Over weight | 4/76 | 1/35 | 0/9 | 1/15 | 0/13 |
| y: Year; SD: Standard Deviation; kg: Kilogram; cm: Centimeter. | |||||
Table 3. Frequency of observed clinical features.
Genotype distributions
Regarding genotyping results, the four selected SNPs (rs35747426, rs121434612, rs199985543 and rs9887672) were incompatible to Hardy-Weinberg equilibrium in both, children with ID and controls. No variation was found in rs35747426 of ARHGEF6 and rs121434612 of PAK3 genes. The frequencies of CC (Leu/Leu), CA (Leu/Pro) and AA (Pro/Pro) genotypes for rs199985543 OPHN1 missense variant among children with ID were 86.20%, 12.9%, and 0.9% respectively. Whereas the frequencies of CC, CT and TT genotypes for rs9887672 IL1RAPL2 intronic variant showed 88.8%, 8.6% and 2.6% respectively (Table 4). The minor allele frequency for ‘A’ of rs199985543 was found to be 0.0001 and for ‘T’ of rs9887672 was found to be 0.18. However, genotype distribution of both the SNPs did not significantly differ in disease and control groups (P=0.35 and P=0.10). Allele distributions (A and T) were significantly different (P=0.005 and P=0.0001). The carrier of genotypes CA (rs199985543) and CT (rs9887672) were significantly different between ID (12.9%; 8.6%) and normal individuals. (P=0.0002 and 0.0023; OR (95% CI)=31.0 (1.82-525.17) and 20.39 (1.17-352.63) respectively). The dominant model analysis also revealed significant association of genotypes CC over CA+AA in OPHN1 gene and CC over CT+TT in IL1RAPL2 gene with intellectual disability risk (P=0.0001 and 0.0005; OR (95% CI)=0.03 (0.0018-0.512) and 0.038 (0.0022-0.650) respectively) (Table 4).
| Gene | SNP ID | Genotype/Allele | Proportion with percent | Statistical analysis | |||
|---|---|---|---|---|---|---|---|
| ID (%) | Controls (%) | P (χ2-test) | P (Fisher exact test) | OR (95% CI) | |||
| OPHN1 | rs199985543 | CC | 100 (86.20) | 100 | |||
| CA | 15 (12.9) | 0 | 0.0002* | 0.0001* | 31.0 (1.82-525.17) | ||
| AA | 1 (0.9) | 0 | 0.3185 | 1 | 3.0 (0.120-74.52) | ||
| AA versus CA+CC | # | # | 0.352 | 1 | 2.61 (0.105-64.80) | ||
| CC versus CA+AA | # | # | 0.0001* | 0.0001* | 0.03 (0.0018-0.512) | ||
| C | 215 | 200 | |||||
| A | 17 | 0 | 0.0052* | 0.0001* | 32.56 (1.94-545.08) | ||
| IL1RAPL2 | rs9887672 | CC | 103 (88.8) | 100 | |||
| CT | 10 (8.6) | 0 | 0.0023* | 0.0019* | 20.39 (1.179-352.63) | ||
| TT | 3 (2.6) | 0 | 0.0901 | 0.247 | 6.79 (0.346-133.27) | ||
| TT versus CT+CC | # | # | 0.1053 | 0.2506 | 6.19 (0.316-121.46) | ||
| CC versus CT+TT | # | # | 0.0005* | 0.0003* | 0.038 (0.0022-0.650) | ||
| C | 216 | 200 | |||||
| T | 16 | 0 | 0.00015* | 0.0001* | 30.56 (1.821-512.76) | ||
| Genotype distributions were shown with number (%), ORs, 95% CI and P values by χ2-test and 2-sided Fisher exact test were calculated from MS Excel spread sheet program and GraphPad online tool; *Indicate significant value; OPHN1: Oligophrenin 1; IL1RAPL2: Interleukin 1 receptor like accessory protein 2; SNP: Single Nucleotide Polyphormism; ID: Intellectual Disability; OR: Odd Ratio; CI: Confidence Interval. | |||||||
Table 4. Association between gene polymorphisms.
Multifactor dimensionality reduction (MDR) analysis
Further analysis of our genotype findings, especially SNP-SNP interaction in children with ID and controls was performed by using Generalized Multifactor Dimensionality Reduction (GMDR) beta 0.9 Software. The parameters include the Cross- Validation Consistency (CVC), the testing accuracy, the training-balance accuracy and the sign test to assess the significance level of SNP model were calculated and summarized in Table 5. The best SNP models accompanied by the lowest prediction error (Testing balance accuracy), the highest CVC and the P value of significant level were calculated. The results revealed the interactions between OPHN1 rs199985543 and IL1RAPL2 rs9887672 as the best SNP model included with testing balance accuracy 64.62% and cross-validation consistency 10/10. The models were significant at 0.001 level.
| Genotype model | Training bal. acc. | Testing bal. acc. | Sign test (p) | CVC |
|---|---|---|---|---|
| IL1RAPL2-rs9887672 | 0.596 | 0.5963 | 8 (0.0547) | 10/10 |
| OPHN1-rs199985543+IL1RAPL2-rs9887672 | 0.6515 | 0.6462 | 10 (0.0010) | 10/10 |
| ARHGEF6-rs35747426+ OPHN1-rs199985543+IL1RAPL2-rs9887672 | 0.6515 | 0.6462 | 10 (0.0010) | 10/10 |
| GMDR: Generalized Multifactor Dimensionality Reduction; SNP: Single Nucleotide Polymorphism; Bal. Acc: Balance Accuracy; CVC: Cross Validation Consistency. | ||||
Table 5. Multifactor dimensionality reduction models.
In silico analysis of OPHN1 protein phenotype prediction
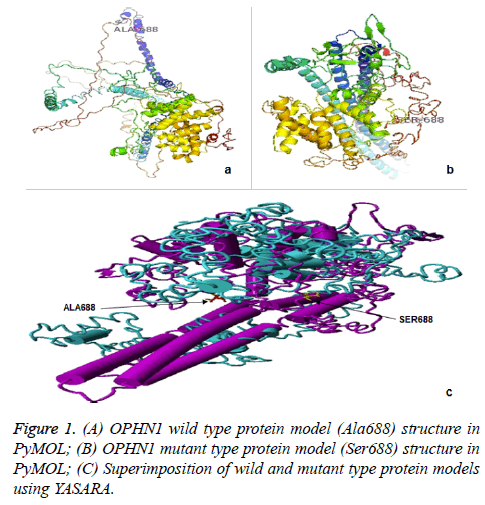
We used 3D structures of native and mutant OPHN1 protein to estimate the structural drift at both, amino acid residue and polypeptide chain levels. RAMPAGE assessment with parameter residues in favoured region (RFR) revealed native model (97.1%) and mutant model (96.9%) have good structural accuracy for downstream structural analysis. The superimposition analysis using YASARA software was resulted in the RMSD value for mutant and native protein structure at amino acid residue (0.59 Å) and polypeptide level (0.61 Å). Similarly, protein stability analysis by DUET and Imutant 2.0 tools, revealed the structure divergence (ΔΔG=-0.011 to -0.318 Kcal/mol). The difference in structures of protein phenotype prediction analysis suggests that OPHN1 rs199985543 polymorphism is damaging and destabilizing (Figure 1).
Discussion
The etiological classification of intellectual disability is under continuous investigations due to the new findings from many genetic studies. More than 700 genes are often found susceptible in the patients with intellectual disability. However, genetic polymorphisms regarding allelic distributions, linkage disequilibrium, and environmental factors often vary across different ethnicities and geographical areas in genetic disorders like intellectual disability. The reported findings demonstrated that ID is involved diversified phenotypic characteristics rather than described with single feature [30]. A genotyping study for celiac disease on Saudi Arabian population was evident potential susceptible locus using bio-statistical analysis [31] and another study on South Indian diabetic patients was signified the result by correlating genotypic and computational approach [32]. Our approach to the current study was based on the necessity to understand the genotype-phenotype consequences and computational protein phenotype predictions in intellectually disabled children of Western Indian population.
The polymorphisms, rs35747426 in ARHGEF6, rs121434612 in PAK3, rs199985543 in OPHN1 and rs9887672 in IL1RAPL2 genes were screened by the standard PCR-RFLP genotyping method in collected samples. The SNPs of PAK3 and IL1RAPL2 genes were previously studied for X-linked intellectual disability [33,34]. Whereas, SNPs of ARHGEF6 and OPHN1 genes were studied for the first time in the current study; from these, three SNPs are missense variant, and one is the intronic variant. In our results, we found a significant statistical difference in allele and genotype frequencies in the case of OPHN1 and IL1RAPL2 genes between diseased and controls. However, it was noted that there was a tendency toward significant associations between carrier genotypes of both the SNPs (P=0.0002) (Table 2).
Oligophrenin 1 (OPHN1) gene encodes a Rho-GTPaseactivating protein (802 amino acids) which is involved in the maintenance of structural and functional integrity in neuronal synapses. The gene is located on chromosome X, and its size spans up to 7879 bp within 25 exons. The genetic polymorphism, rs199985543 is located in exon 21 with the ‘C’ allele codes for alanine and the risk allele ‘A’ codes serine at amino acid position 688. This missense variant is closely located to the Rho-GAP domain of the protein, which is a central functional region of the OPHN1 protein. The phenotypic studies showed the OPHN1 gene mutation is known to cause seizures and cerebral hypoplasia in intellectually disabled children [35].
Interleukin 1 receptor-like accessory protein 2 (IL1RAPL2) gene encodes 686 amino acids long protein of the interleukin 1 receptor family which is closely related to interleukin 1 receptor accessory protein-like 1 (IL1RAPL1). This gene is assigned to chromosome X, and its size spans up to 2985 bp within 11 exons. The intronic polymorphism rs9887672 is located in intron 9 of this gene. The GWAS3D analysis was revealed that this SNP has significant transcription binding affinity, mapping of distal interaction and mapping on the GERP++ conservation element. The data assessed from GWAS Central association studies showed that this mutation at intronic position leads to partial epilepsy and abnormal birth weight. These findings also correlated with epileptic phenotypes present in the 8 affected children with low birth weight in 3 out of whole 13 affected children and they were also found to have IL1RAPL2 genetic variation. Similarly, the children in Qinba region of China were showed the same mutation caused non-syndromic X-linked intellectual disability [34].
ARHGEF6 gene encodes Rho guanine nucleotide exchange factor 6 is the protein (776 amino acids) and PAK3 gene encodes p21-activated kinase 3 protein (803 amino acids). The mutations in both the genes were known to cause XLID with the defects of spine morphogenesis [36]. Leu11Pro is present in ARHGEF6 protein at the N-terminal end of the protein domain of the Calponin homology domain (CHD), and it affects actin binding activity which may lead to intellectual disability with abnormal speech and unusual facial shape. Similarly, Arg67Cys is present on regulatory domain of PAK3 protein at the N-terminal end of the p21-GTPase-binding domain (PBD) and affects kinase activity, which may turn into intellectual disability [37].
Although, ARHGEF6 (Leu11Pro) and PAK3 (Arg67Cys) genes non-synonymous variants were statistically not associated with ID in this study but the bioinformatics data suggested that both genetic mutations are highly deleterious and disease causing variants thus we cannot say that these were not a susceptible loci to cause intellectual disability. Independently, these two loci are also seen under high linkage (by LD scores; r2=1) with genes critical regions involved in disease development in Gujarati-Indians in Houston (GIH), Texas [20].
The phenotypic representation of our data addressed that the majority of clinical features might arise due to the mutations in Rho-GTPases. Studies on Rho-GTPase pathway were demonstrated a fundamental role of Rho-GTPases in numerous cellular processes those extracellular stimuli initiate and that work through G protein coupled receptors [38,39]. The mutations in the Rho-GTPases genes can cause non-specific Xlinked intellectual disability [10].
The GMDR analysis of OPHN1 rs199985543 and IL1RAPL2 rs9887672 polymorphisms has reinforced the significant results that both SNPs increase the risk of developing ID in children.
The present study showed that there was a significant difference in carrier genotype distribution especially rs199985543 in OPHN1 and rs9887672 in IL1RAPL2 gene among children with ID and control group, suggesting that these polymorphisms are susceptible loci to cause intellectual disability. CA genotype for rs199985543 and CT genotype for rs9887672 are the risk factors for ID.
To better understand the function of OPHN1 rs199985543 polymorphism in disease association, it is important to determine the contextual relationship of OPHN1 protein structure and sequence likely to affect. To describe the damaging effect of Ala688Ser variant (rs199985543), 3D structure of mutant OPHN1 protein was modelled and compared RMSD as well as structural stability with native protein. Interestingly, we observed significant deviation in mutant OPHN1 protein structure, both at residue and whole polypeptide levels in case of RMSD and ΔΔG values for protein stability were evident. The RMSD value of identical protein structure is always ‘zero’, and its increase value reflects the difference between two protein structures [40,41]. Similarly, the deleterious variants induce changes in RMSD of amino acid and alter the stability of protein that can be confirmed with negative ΔΔG. In this study, both parameters may confirm that the OPHN1 protein might have damaging effect due to rs199985543 polymorphism.
Sincerely, we are acknowledged following limitations to interpret results in this study. Firstly, we had only 3 missense; 1 intronic genetic polymorphism and did not cover the large numbers of reported SNPs by genotyping studies. Secondly, there is non-availability of gene expression data of the associated genetic markers with linked genetic locations. Similarly, the biology of genetic diseases is much more complex than a directly altered polymorphism associated with disease relationship. A third is the sample size may not be adequate to draw a highly possible conclusion.
Collectively, the study is reported for a first time that OPHN1 rs199985543 and IL1RAPL2 rs9887672 polymorphisms might be a risk factor in causing intellectual disability in the Indian populations of Gujarat state. Our data also showed that the majority of ID affected population having the X-linked phenotypic appearance, but due to the limitation of genotyping study, we cannot identify all the possible genetic mutations. The study may be considered in developing a new hypothesis for the phenotype and genotype research on the Indian population. For further description of these findings, gene expression and other functional analysis are needed.
Ethics Approval
The work has been carried out in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki 1964) for experiments on humans. The Ethical approval for this study was obtained from Human Research Ethics Committee of H. M. Patel Centre for Medical Care and Education, Karamsad, Gujarat, India (Ref. No. HREC/ HMPCMCE/221/15). Additional informed consent was obtained from all individual participants/their parents for whom identifying information is included in this article.
Acknowledgement
We thank all the children and the parents who participated in this study. Authors are thanks to the administrator of the Mamta Hygienic Clinic (Surat), Gurukrupa Residential Special School (Anand), Mitra Rehabilitation Centre (Anand), Deep Special Education for Mental Retardation (Surat) and Deepa Academy (Tarapur) for providing samples. We thank Directors of the ARIBAS and Sophisticated Instrument Centre for Applied Research and Testing (SICART) for providing lab facilities. Financial support provided by Charutar Vidya Mandal (CVM), Vallabh Vidyanagar (partial grant) and University Grants Commission, India (for RGN-Fellowship) thankfully acknowledged.
Conflict of Interest
The authors declare no conflict of interest.
References
- Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. Prevalence of intellectual disability: A meta-analysis of population-based studies. Res Dev Disabil 2011; 32: 419-436.
- Division of Mental Health and Prevention of Substance Abuse World Health Organization. ICD-10 guide for Mental Retardation. Geneva 1996.
- Stevensona RE, Holdena KR, Rogersa RC, Schwartza CE. Seizures and X-linked intellectual disability. Eur J Med Genet 2012; 55: 307-312.
- Kaufman L, Ayub M, Vincent JB. The genetic basis of non-syndromic intellectual disability: A review. J Neurodev Disord 2010; 2: 182-209.
- Raymond FL. X linked mental retardation: a clinical guide. J Med Genet 2006; 43: 193-200.
- Bathelt J, Astle D, Barnes J, Raymond FL, Baker K. Structural brain abnormalities in a single gene disorder associated with epilepsy, language impairment and intellectual disability. Neuroimage 2016; 12: 655-665.
- Vissers LELM, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet 2015; 17: 9-18.
- Merrick J, Morad M, Carmeli E. Intellectual and developmental disabilities: male health. Front Public Heal 2014; 2: 208.
- Schwartz M. Rho signalling at a glance. J Cell Sci 2004; 117: 5457-5458.
- Huang G-H, Sun Z-L, Li H-J, Feng D-F. Rho GTPase-activating proteins: Regulators of Rho GTPase activity in neuronal development and CNS diseases. Mol Cell Neurosci 2017; 80: 18-31.
- Bahi N, Friocourt G, Carrié A, Graham ME, Weiss JL, Chafey P. IL1 receptor accessory protein like, a protein involved in X-linked mental retardation, interacts with neuronal calcium sensor-1 and regulates exocytosis. Hum Mol Genet 2003; 12: 1415-1425.
- Girimaji SC, Srinath S. Perspectives of intellectual disability in India: epidemiology, policy, services for children and adults. Curr Opin Psychiatr 2010; 23: 441-446.
- Khimsuriya Y, Kharod N, Padmani G, Chauhan J, Roy N. Association of an intronic variant of faciogenital dysplasia 1 (FGD1) gene with X-linked intellectual disability. Curr Pediatr Res 2016; 20: 282-287.
- Beck T, Hastings RK, Gollapudi S, Free RC, Brookes AJ. GWAS Central: a comprehensive resource for the comparison and interrogation of genome-wide association studies. Eur J Hum Genet 2014; 22: 949-952.
- Sherry S, Ward M, Kholodov M, Baker J, Phan L, Smigielski E, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001; 29: 308-311.
- Lek M, Karczewski KJ, Eric V, Hill AJ, Cummings BB, Tukiainen T, Donnell-Luria AHO, Ware JS, Tiao G, Tusie-luna MT, Weisburd B, Won H. Analysis of protein-coding genetic variation in 60,706 humans. Nat Publ Gr 2016; 536: 285-291.
- Stenson PD, Mort M, Ball E V, Shaw K, Phillips AD, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet 2014; 133: 1-9.
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res 2003; 31: 3812-3814.
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248-249.
- Li MJ, Wang LY, Xia Z, Sham PC, Wang J, Rr L, May M, Pouls S, Ha L, Re S, Ce S, Teebi AS, Rucquoi JK, Meyn MS. GWAS3D: Detecting human regulatory variants by integrative analysis of genome-wide associations, chromosome interactions and histone modifications. Nucleic Acids Res 2013; 41: 150-158.
- Lahiri DK, Numberger JI. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res 1991; 19: 5444.
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 2002; 12: 996-1006.
- Vincze T, Posfai J, Roberts RJ. NEBcutter: A program to cleave DNA with restriction enzymes. Nucleic Acids Res 2003; 31: 3688-3691.
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat Publ Gr 2015; 12: 7-8.
- Lovell SC, Davis IW, Arendall III WB, Bakker PIW de, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and C beta deviation. Proteins Struct Funct Genet 2002; 50: 437-450.
- Krieger E, Vriend G. Structural bioinformatics YASARA view-molecular graphics for all devices - from smartphones to workstations. Bioinformatics 2014; 30: 2981-2982.
- Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 2005; 33: 306-310.
- Pires DE V, Ascher DB, Blundell TL. DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res 2014; 42: 314-319.
- Motsinger AA, Ritchie MD. Multifactor dimensionality reduction: An analysis strategy for modelling and detecting gene-gene interactions in human genetics and pharmacogenomics studies. Hum Genomics 2006; 2: 318-328.
- Srour M, Shevell M. Genetics and the investigation of developmental delay/intellectual disability. Arch Dis Child 2014; 99: 386-389.
- Saadah OI, Shaik NA, Banaganapalli B, Salama MA, Al-Harthi SE, Wang J, Shawoosh HA, Alghamdi SA, Bin-Taleb YY, Alhussaini BH, Elango R, Al-Aama JY. Replication of GWAS coding SNPs implicates MMEL1 as a potential susceptibility locus among Saudi Arabian celiac disease patients. Dis Markers 2015; 2015: 1-6.
- Rao DK, Murthy DK, Shaik NS, Banaganapalli B, Konda K, Rao HP, Ganti E, Ahmed Awan Z, A El-Harouni A, Elango R, Ali Khan I, Shaik NA. Distribution of CYP2C8 and CYP2C9 amino acid substitution alleles in South Indian diabetes patients: A genotypic and computational protein phenotype study. Clin Exp Pharmacol Physiol 2017; 44: 1171-1179.
- Bienvenu T, Portes, Des V, McDonell N, Carrié A, Zemni R, Couvert P, Ropers HH, Moraine C, van Bokhoven H, Fryns JP, Allen K, Walsh CA, Boué J, Kahn A, Chelly J, Beldjord C. Missense mutation in PAK3, R67C, causes X-linked nonspecific mental retardation. Am J Med Genet 2000; 93: 294-298.
- Zhang KJ, He B, Gong PY, Gao XC, Zheng ZJ, Huang SP, Zhang FC. An association study between IL1RAPL2 gene and non-specific mental retardation in Chinese children. Genes and Genomics 2010; 32: 159-164.
- Zanni G, Saillour Y, Nagara M, Billuart P, Castelnau L, Moraine C, Faivre L, Bertini E, Durr A, Guichet A, Rodriguez D, Des Portes V, Beldjord C, Chelly J. Oligophrenin 1 mutations frequently cause X-linked mental retardation with cerebellar hypoplasia. Neurology 2005; 65: 1364-1369.
- Nodé-Langlois R, Muller D, Boda B. Sequential implication of the mental retardation proteins ARHGEF6 and PAK3 in spine morphogenesis. J Cell Sci 2006; 119: 4986-4993.
- Kreis P, The´venot E, Rousseau V, Boda B, Muller D, Barnier J-V. The p21-activated Kinase 3 implicated in mental retardation regulates spine morphogenesis through a Cdc42-dependent pathway. J Biol Chem 2007; 282: 21497-2506.
- Ba W, Raadt J van der, Kasri NN. Rho GTPase signaling at the synapse : Implications for intellectual disability. Exp Cell Res 2013; 319: 2368-2374.
- Elly JM, Van G, Ramakers GJA. Rho proteins, mental retardation and the neurobiological basis of intelligence. In: Progress in Brain Res 2005; 295-317.
- Carugo O, Pongor S. A normalized root-mean-square distance for comparing protein three-dimensional structures. Protein Sci 2008; 10: 1470-1473.
- Maiorov VN, Crippen GM. Significance of root-mean-square deviation in comparing three-dimensional structures of globular proteins. J Mol Bio 1994; 235: 625-634.