ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 11
Extraction of breast border and removal of pectoral muscle in wavelet domain
Bushra Mughal1, Nazeer Muhammad2*, Muhammad Sharif1, Tanzila Saba3 and Amjad Rehman4
1Department of Computer Science, COMSATS Institute of Information Technology, Wah Cantt, Pakistan
2Department of Mathematics, COMSATS Institute of Information Technology, Wah Cantt, Pakistan
3Department of Information Systems, Prince Sultan University in Riyadh, Saudi Arabia
4College of Computer and Information Systems, Al-Yamamah University, Riyadh, Saudi Arabia
- *Corresponding Author:
- Nazeer Muhammad
Department of Mathematics
COMSATS Institute of Information Technology, Pakistan
Accepted on April 1, 2017
Extraction of the breast border and simultaneously exclusion of pectoral muscle are principal steps for diagnosing of breast cancer based on mammogram data. The objective of propose method is to classify the mediolateral oblique fragment of the pectoral muscle. The extraction of breast region is performed using the multilevel wavelet decomposition of mammogram images. Moreover, artifact suppression and pectoral muscle detection is carried out by morphological operator. The efficient extraction with higher accuracy is validated on the set of 322 digital images taken from MIAS dataset.
Keywords
Pectoral muscle, Mediolateral oblique, Cranial-Caudal, Computer-aided detection system
Introduction
Breast cancer is fatal disease of females almost worldwide and every year death rate is increased gradually due to its metastatic sites [1]. Computer-Aided Detection (CAD) setup may establish a significant contribution in the early age detection of tumor cell in women’s breast and may improve their mortality rate [2].
A novel technique is presented in [3] for removal of pectoral muscle [4]. Cartoon texture is introduced for localization of pectoral muscle [5]. A breast border line detection method based on the contours of homogeneous regions is presented [6,7]. Combination of morphological operators and polynomial function are introduced for pectoral muscles detection in [8]. Various methods are presented in literature for removal of pectoral muscle in [9,10]. Graph theory is used in combination of active contours for removing of pectoral muscle [11]. Bezier curve is used for smoothing the pectoral muscle by tracking their edge points [12]. A normalized graph cuts based method is designed for isolating the pectoral muscle edge [13]. A muscle contour detection method based on shortest path is presented [14]. Bit depth reduction and wavelet transform is used for pectoral muscle removal [15]. Mostly pectoral muscle appears in mammogram which are obtained from Mediolateral- Oblique (MLO) fragment. Pectoral muscle is considered a dominant section of mammograms that contain significant information and also affects the segmentation, feature extraction and classification process which causes the high rate of false positive, so it is needed to remove pectoral muscle.
Due to the variations in size, shapes, intensity decomposition, it becomes very difficult to isolate pectoral muscle from breast border line. In this regard, we propose extraction of breast border using mathematical morphology in wavelet domain.
Our paper is defined as follows: In Section 3, proposed procedure and its graphical representation is illustrated. In Section 4, performance assessment is accomplished. In Section 5, we conclude our work.
Proposed Methodology
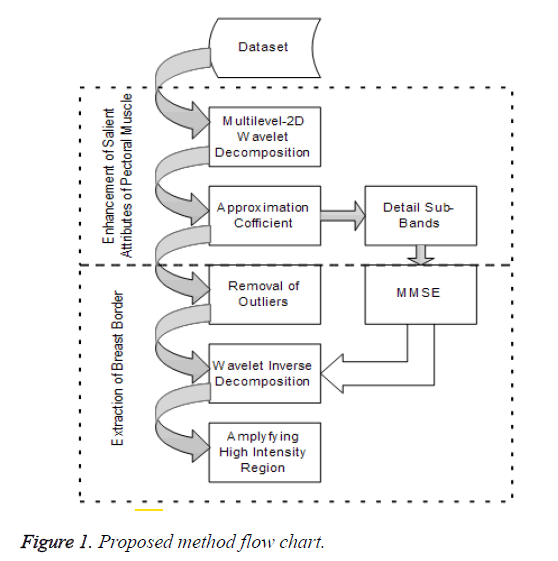
Most of the existing methods to detect the pectoral muscles are providing less accurate results, usually, mislead the information in case of tumor segmentation accuracy. Therefore, it is required to improve the existing methods for attaining high level of accuracy. A sequential flow of the proposed method is shown in Figure 1.
Removal of pectoral muscle
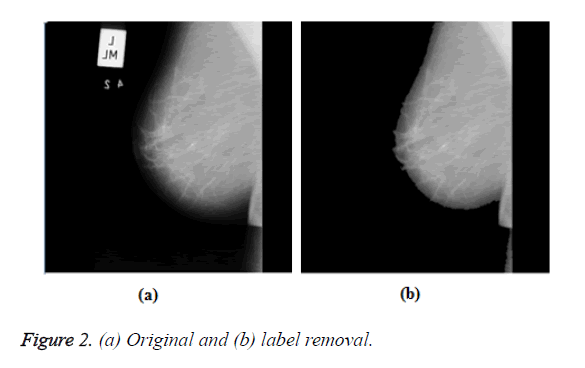
Mammographic images have two parts, one is the visible breast area and the other is background area that may contain radiopaque artifacts, markers, and labels that can be removed by amplifying the high intensity area using a seed point approach. A mammogram before and after removal of these artifacts is displayed in Figure 2.
Median filter is used for removing the quantum noise present in mammographic images. Most of the heuristic based noise removal filters blur the given image [16]. Therefore, we use mathematical formulation of the most adopted filter as described in Equation 1.
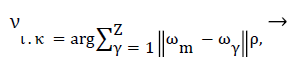 (1)
(1)
Where υικ is the output value for location xικ, νικ=ν1, ν2, ν3, ….νΖ, ι,κ are representing the position in a 2D image, m and γ are filtering window indices, where filtering window length is denoted by Z, and ρ denote the Lρ norm. Commonly, ρ=1 corresponds to standard median filtering. The main purpose of this preprocessing is to obtain significant mammograms that further help us to obtain accurate breast profile. We proposed a unique method to for removal of pectoral muscle in order to achieve accurate tumor region.
Enhancement of salient feature of pectoral muscle
Pectoral muscle appears as intensity region on mammograms having no significant information about tumor recognition and may cause erroneous pathological decisions. So, removal of pectoral muscle from breast parenchyma is considered important properties of computer aided diagnoses system for accurate tumor segmentation and classification. In this proposed work, filtered mammograms are decomposed in four sub band by using multilevel 2D wavelet decomposition [3,17,18]. One band is approximation scale and other three bands are detailed sub bands. The approximation scale hold out the approximation coefficients and the other three bands preserve horizontal, vertical and diagonal details, respectively. Mathematical functions of wavelet transform can be given as follows
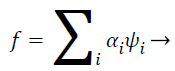 (2)
(2)
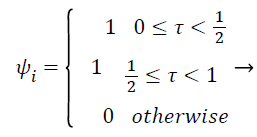 (3)
(3)
Extraction of breast border line
For refining the breast parenchyma from outlier’s contour operation in combination with morphological operators is applied on approximation sub band. Minimum mean squared error (MMSE) [19] is applied on detailed sub bands. Inverse Wavelet decomposition is applied on all four sub bands and amplifying the areas of high intensity and segment them using a seed. The seed is adjusted on the convex hull and erodes the map until it has touched on the edge of the areas to keep, preserving the geometry of breast region. The generation of wavelet coefficients based on two levels is called a dyadic scale which leads to get an efficient and accurate analysis [2,3]. The analysis of any signal β provides the results in form of coefficients Ρ (ω, Υ) as follows:
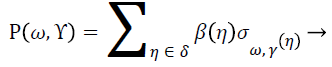 (4)
(4)
Experiments and Results
Let a mammogram be represented by the pixel set ω={ω1, ω2…, ωn} with |ω|=row × col of the matric on which the image is defined. The performance is evaluated on total 322 mammograms (μ) based on MIAS database. In subjective method, visual check by an experienced radiologist is often adopted to measure the performance evaluation of proposed work (Table 1).
| Category | Evaluation criteria | No of images | Proposed |
|---|---|---|---|
| Accurate (α) | 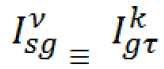 |
250 | 77.63% |
| Acceptable (ρ) | 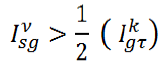 |
47 | 14.59% |
| Unacceptable (γ) | 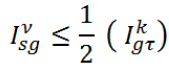 |
25 | 7.76% |
| Overall Accuracy (Αϖ) | 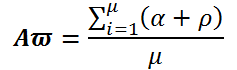 |
322 | 92.22% |
Table 1. Evaluation of proposed segmentation technique.
Conclusion
We propose moderate segregation of the pectoral muscle in well-known dataset of MIAS images. For analysis purpose of mammogram images, the required pectoral muscle boundaries drawn on manual basis by an expert radiologist. For validation and comparison, we use these manual drawn boundaries images as a ground truth. The test results of the proposed method are promising and very much useful for primary level detection of breast cancer.
Acknowledgements
We would like to thank Mr. Ayaz (radiologist) from St. Jude Children's Research Hospital, Memphis, US for providing us the ground truth data for segmenting the breast mass to evaluate the improvement of our proposed method.
References
- Pobiruchin M, Bochum S, Martens UM, Kieser M, Schramm W. A method for using real world data in breast cancer modeling. J Biomed Inform 2016; 60: 385-394.
- Tang J, Rangayyan RM, Xu J, El Naqa I, Yang Y. Computer-aided detection and diagnosis of breast cancer with mammography: recent advances. IEEE Trans Inform Technol Biomed 2009; 13: 236-251.
- Mustra M, Bozek J, Grgic M. Breast border extraction and pectoral muscle detection using wavelet decomposition. Breast Border Extraction Pectoral Muscle Detection Using Wavelet Decomposition IEEE 2009; 1426-1433.
- Firmino M, Angelo G, Morais H, Dantas MR, Valentim R. Computer-aided detection (CADe) and diagnosis (CADx) system for lung cancer with likelihood of malignancy. Biomed Eng 2016; 15: 1.
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J. Global cancer statistics, 2012. CA Cancer J Clin 2015; 65: 87-108.
- Lakshmanan, R, Thomas, V, Jacob, S.M, Thara, P. Pectoral Muscle Boundary Detection in Mammograms Using Homogeneous Contours. Book Pectoral Muscle Boundary Detection in Mammograms Using Homogeneous Contours IEEE 2015; 354-357.
- Ferrari RJ, Rangayyan RM, Desautels JL, Borges R, Frere AF. Automatic identification of the pectoral muscle in mammograms. IEEE Trans Med Imag 2004; 23: 232-245.
- de Carvalho IM, Luz L, Alvarenga A, Infantosi A, Pereira W, Azevedo C. An automatic method for delineating the pectoral muscle in mammograms. Book An Automatic Method for Delineating the Pectoral Muscle in Mammograms Springer 2007: 271-275.
- Vidivelli S, Devi SS. Breast region extraction and pectoral removal by pixel constancy constraint approach in mammograms. Comp Intell Cyber Secur Comp Models Springer 2016: 195-206.
- Wang L, Zhu M, Deng LP, Yuan X. Automatic pectoral muscle boundary detection in mammograms based on Markov chain and active contour model. J Zhejiang Univ Sci C 2010; 11: 111-118.
- Ma F, Bajger M, Slavotinek JP, Bottema MJ. Two graph theory based methods for identifying the pectoral muscle in mammograms. Patt Recogn 2007; 40: 2592-2602.
- Camilus KS, Govindan VK, Sathidevi PS. Computer-aided identification of the pectoral muscle in digitized mammograms. J Digit Imaging 2010; 23: 562-580.
- Abdellatif H, Taha T, Zahran O, Al-Nauimy W, El-Samie FAK. Automatic pectoral muscle boundary detection in mammograms using eigenvectors segmentation. Automatic Pectoral Muscle Boundary Detection in Mammograms using Eigenvectors Segmentation IEEE 2012; 633-640.
- Domingues I, Cardoso JS, Amaral I, Moreira I, Passarinho P, Santa Comba J, Correia R, Cardoso MJ. Pectoral muscle detection in mammograms based on the shortest path with endpoints learnt by SVMs. Pectoral Muscle Detection in Mammograms Based on the Shortest Path with Endpoints Learnt by SVMs IEEE 2010; 3158-3161.
- Rouhi R, Jafari M, Kasaei S, Keshavarzian P. Benign and malignant breast tumors classification based on region growing and CNN segmentation. Exp Sys Appl 2015; 42: 990-1002.
- Gan S, Wang S, Chen Y, Chen X, Xiang K. Separation of simultaneous sources using a structural-oriented median filter in the flattened dimension. Comp Geosci 2016; 86: 46-54.
- Muhammad N, Bibi N. Digital image watermarking using partial pivoting lower and upper triangular decomposition into the wavelet domain. IET Image Proc 2015; 9: 795-803.
- Raviraj P, Sanavullah M. The modified 2D-Haar Wavelet Transformation in image compression. Middle East J Sci Res 2007; 2: 73-78.
- Tuchler M, Singer AC, Koetter R. Minimum mean squared error equalization using a priori information. IEEE Trans Sig Proc 2002; 50: 673-683.