ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 10
Expression of the matrix gla protein (MGP) gene in different developmental stages of mouse mandibular first molar root
1Research Institute of Stomatology, General Hospital of PLA, Beijing, PR China
2Department of Stomatology, the 306th Hospital of PLA, Beijing, PR China
#These authors contributed equally to this work
- *Corresponding Author:
- Zhongying Niu
Department of Stomatology
The 306th Hospital of PLA, Beijing, PR China
Accepted on February 22, 2017
The objective of the study was to analyse the expression of the matrix Gla protein gene, MGP, in different developmental stages of mouse mandibular first molar root. Laser microdissection was used to cut dental follicular tissues of the first molar in mouse embryos at 17 days of development (E17) and two days after birth (PN2). Total RNA of the dental follicular tissues was extracted to prepare a microarray for screening the MGP gene expression at E17 and PN2, and results were verified using the Polymerase Chain Reaction (PCR) method. The microarray results from the tooth germ tissue taken from mice at different developmental stages showed that MGP gene expression in tissue taken at E17 was higher than that in tissue taken at PN2. PCR results confirmed the microarray results (P<0.05). This study examined the expression of the MGP gene in mouse germ tissues obtained by laser microdissection at different times during development (pre-natal and post-natal). These data show that the MGP gene expression decreased after birth, indicating a role of the MGP gene in development of the tooth root.
Keywords
Gene chip, Mgp gene, Mouse tooth germ, Root development.
Introduction
The Matrix Gla Protein (MGP) is a member of the Extracellular Matrix (ECM) family of proteins that play an important role in the development of tooth and bone and in the calcification of the tooth extracellular matrix, and are regulated by the activity of metal ions, proteins, and other cytokines [1-5]. The vitamin K-dependent MGP gene was initially discovered in cow bones and was subsequently discovered to exist in the teeth, heart, lung, cartilage, and other tissues [3,6-8]. It was not known whether the MGP gene was involved in the process and mechanism of tooth germ development. In the mouse MGP gene-deficient model, calcified plaques were found in mouse vital arteries, which affected vascular elasticity leading to blood vessel rupture [9]. Non-functioning of the MGP gene can cause calcification of the extracellular matrix and promote cartilage calcification. These studies indicated that the MGP gene was an inhibitor of vascular and cartilage calcification and played a role in the prevention of atherosclerosis and cartilage calcification [10]. In this experiment, the MGP gene/expression profiles of first molar mandibular follicle tissues of CD1 mice at embryonic day 17 (E17) and two days after birth (PN2) were analysed and compared using the enrichment analysis software DAVID [11]. Analysis of the MGP gene was performed to determine whether it was involved in the process of tooth germ tissue development and whether and expression of the gene changed at different developmental stages of tooth root development in CD1 mice. The aims of the study were to lay the foundation for further studying the mechanism of the MGP gene in tooth germ development in CD1 mouse, and to add to the database of the genes involved in tooth developmental regulation.
Materials and Methods
Animals
CD1 mice were provided by Vital River Experimental Animal Technical Co., LTD (Beijing). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol used has been reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the 306th Hospital of PLA (Chinese People's Liberation Army).
Sampling
Mouse mandibular tissues were taken during embryonic 17 day (E17) and two days after birth (PN2) in accordance with the experimental time points and were rinsed in DEPC-PBS (Invitrogen, USA). The centrifuge tubes containing the samples were filled with isopentane (Sigma, USA), put into a liquid nitrogen box for 6-8 s, taken out, transferred into Optimal Cutting Temperature (OCT) embedding medium (Shandon, UK), and finally stored in liquid nitrogen.
Preparation of frozen sections
The embedded tissue blocks were roughly cut into 40 μm sections after roughly visualizing the tooth form and were then cut into two 20-25 μm fine sections. Tooth germs and dental follicular tissues were chosen under a light microscope, covered with Laser Metal Deposition (LMD) coating, sliced and collected. Ten slices of 20-25 μm thickness were collected. The coated tissue sections were pre-treated with 100% ethanol (Merck KGaA, Germany) for 5-10 s and after drying, were reserved for laser microdissection (LMD USA).
Laser microdissection
Leica microdissection operating software (Leica, Germany) was used with setup parameters of: magnification of 10X-20X, continuous closed line, accurate cutting precision, and cutting time. The sections were placed in a slide bracket, 0.2 ml Polymerase Chain Reaction (PCR) tube was placed on the pipe holding rack, 30 μL cell lysates (Invitrogen, USA) was added into the tube cap; started cutting after selecting target tissue or cell populations, PEM membrane with selected target cell population fell into cell lysis buffer in the tube cap [12].
Extraction, amplification of RNA and cRNA purification
The modified trizol method was used to extract RNA organization. The procedure was performed in accordance with the instructions of RNA fluorescent linear amplification kit (Agilent, USA) and cRNA purification kit (Agilent, USA).
Microarray hybridization
2X cRNA sample solution: 0.75 μg Cy3-labeled linear amplification cRNA, 0.75 μg Cy5-labeled linear amplification cRNA, 50 μl 10X control quality control samples (control target), adjusted by nuclease-free water to a final volume of 215 μl. 2X hybridization solution: 9.0 μl of 25X fragmentation buffer was mixed with 2X cRNA sample solution. The samples were incubated iPCRn the dark at 60°C for 30 min. 225 μl of 2X hybridization buffer solution was added and mixed.
Hybridization: Agilent coverslip kit was put into hybridization box, 400 μL of 2X hybridization solution was added on the coverslip, put Agilent chip into the hybridization box, hybridized for 17h at 60°C, 4 rpm.
Analysis of microarray raw data
The prepared samples were read in the Agilent microarray scanner to obtain hybridization signal data. Quantitative conversion of the data was performed using the signal extraction software from Agilent. All data were normalized, and two-fold differentially expressed gene selection was done by Agilent GeneSpringGX 7.3 software.
Gene enrichment analysis
Gene enrichment analysis was performed using the DAVID site (http://david.abcc.ncifc/gov/home). The “functional annotation” was selected in the analysis interface and gene lists were uploaded. Next, the Agilent probe identifier number was selected, the list type was set to the uploaded gene lists and the job was submitted and analysed.
RT-PCR
The cDNA reverse transcription reaction contained the following components: RNA, 2 μl; 50 μM oligo (dT), 1 μl; 10 mM dNTP mix, 1 μl; DEPC-treated water, 6 μl, reacted for 5 minutes at 65°C and then placed on ice. The cDNA amplification system contained the following components: 10X RT buffer, 2 μl; 25 mmol/L Mgcl2, 4 μl; 0.1 M DTT, 2 μl; RNaseOUT (40 U/μl), 1 μl; SuperScript III RT (200 U/μl), 1 μl. The 10 μl mixture described previously was added to the reaction mixture that had been placed on ice. The sample was next subjected to the following reaction conditions: 50°C for 50 min, 85°C for 5 min, placed on ice, added 1 μl of RNase H, and 37°C for 20 min. The final cDNA product was stored at -20°C. PCR primers used in the generation of the cDNA are shown in Table 1.
| Gene | Primer sequences | Annealing temperature | Length of products |
|---|---|---|---|
| GAPDH | 5’TCCACCACCCTGTTGCTGTA 3’ | 60 | 450 bp |
| 5’ACCACAGTCCATGCCATCAC 3’ | |||
| Mgp | 5’ ACCCGAGACACCATGAAGAG 3’ | 48 | 240 bp |
Table 1. Primer sequences, annealing temperature and the length of products for target gene and reference gene GAPDH.
Statistical analysis
Each target gene sample analysed in the semi-quantitative PCR experiments was repeated three times. If the repeated results were consistent, then the mean and standard deviation were obtained. If one of the results was not consistent, this value was discarded and the mean and standard deviation of the remaining two samples were obtained. The SPSS16.0 package software was used to perform statistical analysis of the RNA sample integrity values (RNA Integrity Numbers or RIN values) between the study groups and the control 28S/18S rRNA ratio to evaluate the mean and standard deviations. The difference among groups was analysed using multiple analyses of variance, and pairwise comparison between groups was carried out using Student-Newman-Keuls test.
Results
Mouse tooth germ tissues
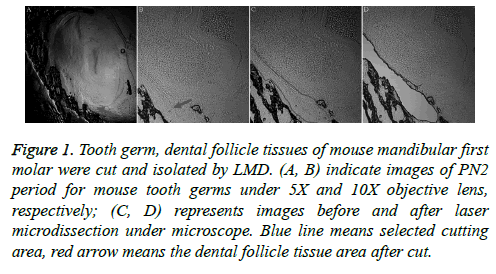
Laser microdissection is a more precise method, and it was used to cut tooth germ and dental follicle tissues of mouse mandibular first molars (Figure 1). The dental follicular tissues surround the outer periphery of the tooth enamel organ. The ectodermal mesenchymal tissues are arranged in a ring surrounding the enamel organ and the bottom of the dental papilla. A clear form of dental papilla is shown in Figure 1A. The pre-cut dental follicular tissue is located on the bottom of dental papilla (Figures 1A and 1B).
Figure 1: Tooth germ, dental follicle tissues of mouse mandibular first molar were cut and isolated by LMD. (A, B) indicate images of PN2 period for mouse tooth germs under 5X and 10X objective lens, respectively; (C, D) represents images before and after laser microdissection under microscope. Blue line means selected cutting area, red arrow means the dental follicle tissue area after cut.
Quality control of RNA samples
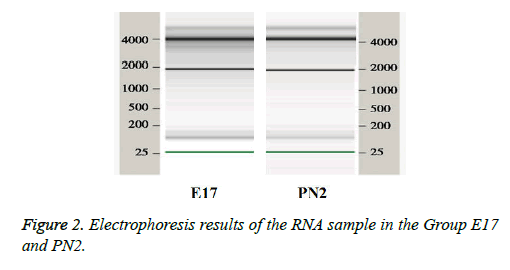
The quality control of RNA extracted from tissue samples was performed on RNA 6000 microfluidic chips used in the 2100 Bioanalyzer from Agilent (United States). The ribosomal RNA 28S/18S ratio (28S/18S Ratio) and the RNA integrity number (RIN) were used as standards. The 28S/18S Ratio in E17 group was 2.01 and the RIN was 7.2. The 28S/18S Ratio in PN2 group was 2.05 and the RIN was 7.9 indicating that RNA was pure. The 18S and 28S RNA bands can be clearly seen in the sample gel electrophoresis picture (Figure 2), suggesting that RNA was not contaminated and can be used in microarray experiments.
Microarray bioinformatics analysis
The mouse whole genome chip purchased from Agilent (United States) contains more than 42,000 probe sequences. The E17 and PN2 dental follicular tissues examined in this study were compared, and 36000 credible gene results were obtained. The two sets of microarray data were compared and analysed to determine the differential expression of the MGP gene in the tissue samples. The criterion used for the expressed genes was a ratio of ≥ 2.
The two-fold upregulated and downregulated genes in the results obtained from the mouse whole genome chip analysis were input to the DAVID online platform, the results showed several enriched genes were gotten.
Gene ontology (GO) analysis
As shown in Table 2 the MGP gene was one of the genes down-regulated more than two-fold in the comparison of the tissues from the E17 and PN2 time periods using the subdirectories of extracellular matrix sequences.
| Genebank accession | Gene name |
|---|---|
| NM_199473 | Collagen, type VIII, alpha 2 |
| AK052759 | Decorin |
| NM_007925 | Elastin |
| NM_181849 | Fibrinogen beta chain |
| NM_008597 | Matrix gla protein |
| NM_008695 | Nidogen 2 |
| NM_198112 | Osteocrin |
| NM_012050 | Osteomodulin |
Table 2. Comparisons of dental follicle tissues during PN2 and E17 two periods under subdirectories of ECM function. Gene list of the top eight ranked 2 times down-regulated expression genes.
RT-PCR
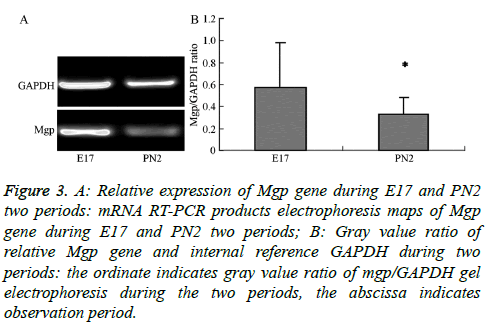
The electropherogram of PCR products obtained for the MGP gene in the samples from the E17 and PN2 time-periods is shown in Figure 3A. The ratios of the intensity of the target gene to the reference gene, Glceraldehyde-3-Phosphate Dehydrogenase (GAPDH), were measured by gel image analyser (Figure 3B). This ratio was obtained as a correction factor (Table 3) to compare with the normalized value of the gene expression obtained in the chip experiment for tissues from the E17 and PN2 periods to verify expression trends. The results show that MGP gene microarray validated results were consistent with the observed trends in the E17 and PN2 periods confirming that the MGP gene expression was higher during the E17 time period compared to expression during the PN2 time period.
Figure 3: A: Relative expression of Mgp gene during E17 and PN2 two periods: mRNA RT-PCR products electrophoresis maps of Mgp gene during E17 and PN2 two periods; B: Gray value ratio of relative Mgp gene and internal reference GAPDH during two periods: the ordinate indicates gray value ratio of mgp/GAPDH gel electrophoresis during the two periods, the abscissa indicates observation period.
| Mgp microarray normalized values | 1 | 0.38 |
| Mgp/GAPDH gray value ratio | 0.57 ± 0.41 | 0.32 ± 0.15 |
Table 3. Experiments normalized values of Mgp microarray and mgp/ GAPDH gray value ratio in RT-PCR reaction.
Discussion
Eert et al. [12] from the US National Institutes of Health developed Laser Capture Microdissection (LCM), which enabled convenient and precise isolation of a single cell or group of cells from native tissue in a complete form for examination under a powerful microscope [13]. Hoffmann et al. [14] obtained relatively pure mouse dental papilla, dental pulp cells, odontoblasts, cementoblasts, and periodontal ligament fibroblasts of mandibular first molar by LCM. Some scholars performed differential protein analysis of mesenchymal cells during early tooth development using the LCM technology combined with two-way gel electrophoresis and mass spectrometry analysis [15,16].
In this study, dental sac’s development derived from mesenchymal tissue of the embryonic neural crest origin. In the 15 days of mouse embryos period (E15), also namely tooth germ development went into the cap stage, ecto-mesenchymal cells divided into two kinds of cells: one is the future form of odontoblasts and pulp cells, dental papilla cells eventually become the dentine-pulp complex; Another is for future differentiation of bone cells, periodontal membrane cells into osteoblast and dental sac cells. From the morphological point, 17 d (E17, embryonic period), dental papilla cells divided into odontoblasts, and the root development would be end after the birth. What factors lead to the different differentiation between two tissues originated from the neural crest tissue from E17? In the early stage of root development, if there had genes differential expression sooner than the morphological changes lead to different differentiation of these two tissues? In the different stage of the same tissue, which genes differential expression leads to tissue specificity? In order to answer these questions, we selected the E17 and PN2, used LMD cut specific dental sac tissue, lay the foundation for subsequent difference genetic screening. Dental follicle tissues were isolated from mouse mandibular first molar at two different time periods (E17 and PN2) to make frozen sections and perform post-production laser cutting. This method can accurately get samples which experiments needed. The results in this study show that laser microdissection technology can enhance the cutting accuracy and efficiency of dental follicle tissues, provide RNA sample of high quality for making gene chips, and lay the foundation for subsequently comparing differential gene expression in different phases of the tissue development and differentiation process using a highthroughput platform.
The advantages of using microarray experiments are automation and high-throughput. The labeling, hybridization and other processes of many gene probes can be done in a single experiment, and the results are automatically analysed by software, which was fast and convenient. But the method is also susceptible to a variety of pitfalls including a high rate of false-positives. To validate results from microarray experiments, Northern blot or in situ hybridization (in-situ) or other methods are needed to verify the differential expression of the screened genes. PCR technology is one of the most simple and effective experimental means for verification of microarray results. The results of this study were verified by PCR, suggesting that the trends of MGP gene expression in the E17 and PN2 time periods were consistent were consistent with the microarray data showing a decrease in expression during the E17 to PN2 timeframe. This finding reminder us to find if MGP gene participate in mediation tooth germ differentiation. Under normal circumstances differentiation of precursor cells in the regeneration process are triggered in instances of tooth root development or tissue repair by a cellular “micro-environment” (niche), which consists of Extracellular Matrix (ECM) and matrix growth factors [5,17]. ECM is a multi-dimensional structure that contains collagen, fibronectin, elastin, and other non-collagenous proteins. Its main function is promoting cell adhesion, cell shape extension, tissue expansion, and cytoskeleton formation. A type of “stem cell” secreted and synthesized by the extracellular matrix may play important roles in altering local microenvironment, and triggering differentiation of the next generation "stem cell" [18]. Thus, more and more studies are examining the gene expression and distribution of ECM family. The MGP gene is associated with typical function in the extracellular matrix and has synergism with osteocalcin (OC) [19]. Research performed in other laboratories used a 2.3 kb central promoter to expressed the MGP gene in osteoblasts, odontoblasts and cementoblasts of transgenic mice and showed that bone mineralization was inhibited [20,21]. In Colla1-MGP transgenic mice, it was found that mineralization appeared to be missing in the root dentin and cementum, and was accompanied by a decrease in alveolar bone density [22,23]. Clinically, patients with osteoporosis, usually with MGP gene activation. Many scholars regard the MGP gene as the purpose therapy gene of osteoporosis; thought that it was involved in the pathogenesis of osteoporosis, especially in postmenopausal osteoporosis disease, mgp expression was activated. Mgp was also be an important regulatory factor of endochondral bone formation and vascular calcification, was a kind of important mineral salt deposition inhibitor, its abnormal expression closely related to extracellular matrix abnormal calcification of tissues such as cartilage and artery [24,25]. In this experiment, the results showed that MGP gene expression at two days after birth was significantly decreased compared with that on embryonic 17. The results of this study are consistent with the results of Murshed et al. [6], who suggested that MGP inhibited bone mineralization and reduced bone density. This change may be related with continuous development of mouse tooth germ after birth, and the increase in the degree of mineralization; that is, MGP expression is reduced and accompanied with an increase in the degree of mineralization of the tooth. This phenomenon may be related to differences between the external environment after the birth of the embryo and the embryonic internal environment resulting in the change of MGP gene expression.
In summary, the development of tooth germ tissue is completed in a unified regulation of a complex molecular regulatory network. The expression of the MGP gene was examined in dental tissues taken from mouse embryos (E17) and compared with dental tissues taken after birth (PN2). The change may be related to increased mineralization of developing tooth germ after mice was born. The results of this experiment showed that the MGP gene might play a role in the developmental process of mouse tooth germ tissues involved in tooth development in mice. Also this result may relate to the internal environment differences between embryonic period and after born, which leading to the change of MGP gene’s expression. Further studies will be needed to understand the regulation mechanism and variation in expression of the MGP gene, add group for exact division of time to find more ways and gene mediating tooth development.
Acknowledgements
This study was supported by the State key laboratory of aerospace medicine foundation and application of open issue (Differentiation gene research of periodontal ligament osteoblastic differentiation under simulated weightlessness based on microarray data screening of rabbits, SMFA14K03) and the Capital health development research project of Beijing, China (Grant No.2014-4-5051).
Conflict of Interest
All authors have no conflict of interest regarding this paper.
References
- Scheller EL, Krebsbach PH, Kohn DH. Tissue engineering: state of the art in oral rehabilitation. J Oral Rehabil 2009; 36: 368-389.
- Jussila M, Thesleff I. Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harb Perspect Biol 2012; 4: a008425.
- Sasaki H, Muramatsu T, Kwon HJ, Yamamoto H, Hashimoto S. Down-regulated genes in mouse dental papillae and pulp. J Dent Res 2010; 89: 679-683.
- Addison WN, Azari F, Sorensen ES, Kaartinen MT, McKee MD. Pyrophosphate inhibits mineralization of osteoblast cultures by binding to mineral, up-regulating osteopontin, and inhibiting alkaline phosphatase activity. J Biol Chem 2007; 282: 15872-15883.
- Ma Y, Ji Y, Huang G, Ling K, Zhang X, Xu F. Bioprinting 3D cell-laden hydrogel microarray for screening human periodontal ligament stem cell response to extracellular matrix. Biofabrication 2015; 7: 044105.
- Murshed M, Harmey D, Millan JL, McKee MD, Karsenty G. Unique co-expression in osteoblasts of broadly expressed genes accounts for the spatial restriction of ECM mineralization to bone. Genes Dev 2005; 19: 1093-1104.
- Li X, Zhou M, Wang X, Li R, Han N, Zhang Q. Quantitative determination of high-temperature requirement protein A1 and its possible associated molecules during induced reparative dentin formation. J Endod 2012; 38: 814-820.
- Ripamonti U. Recapitulating development: a template for periodontal tissue engineering. Tissue Eng 2007; 13: 51-71.
- Date Y, Yokoyama Y, Kondo H, Kuroda S, Ohya K, Ota MS, Iseki S, Kasugai S. Restricted expression of chromatin remodeling associated factor Chd3 during tooth root development. J Periodontal Res 2012; 47: 180-187.
- Marulanda J, Gao C, Roman H, Henderson JE, Murshed M. Prevention of arterial calcification corrects the low bone mass phenotype in MGP-deficient mice. Bone 2013; 57: 499-508.
- Huangda W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009; 37: 1-13.
- Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. J Science 1996; 274: 998-1001.
- Moco S, Schneider B, Vervoort J. Plant micrometabolomics: the analysis of endogenous metabolites present in a plant cell or tissue. J Proteome Res 2009; 8: 1694-1703.
- Hoffmann M, Olson K, Cavender A, Pasqualini R, Gaikwad J. Gene expression in a pure population of odontoblasts isolated by laser-capture microdissection. J Dent Res 2001; 80: 1963-1967.
- Bornstein SR, Willenberg HS, Scherbaum WA. Progress in molecular medicine: laser capture microdissection. Med Klin (Munich) 1998; 93: 739-743.
- Clement-Ziza M, Munnich A, Lyonnet S, Jaubert F, Besmond C. Stabilization of RNA during laser capture microdissection by performing experiments under argon atmosphere or using ethanol as a solvent in staining solutions. RNA 2008; 14: 2698-2704.
- Zhang YD, Chen Z, Song YQ, Liu C, Chen YP. Making a tooth: growth factors, transcription factors, and stem cells. Cell Res 2005; 15: 301-316.
- Wu RX, Bi CS, Yu Y, Zhang LL, Chen FM. Age-related decline in the matrix contents and functional properties of human periodontal ligament stem cell sheets. Acta Biomater 2015; 22: 70-82.
- Kaipatur NR, Murshed M, McKee MD. Matrix Gla protein inhibition of tooth mineralization. J Dent Res 2008; 87: 839-844.
- Li R, Li X, Zhou M, Han N, Zhang Q. Quantitative determination of matrix Gla protein (MGP) and BMP-2 during the osteogenic differentiation of human periodontal ligament cells. Arch Oral Biol 2012; 57: 1408-1417.
- Li DH, Fu HD, Liu WL, Wei YS, Jiang DM. Decreased local and systematic matrix Gla protein (MGP) expression and its link to radiographic progression in ankylosing spondylitis patients. Clin Lab 2015; 61: 243-249.
- Malhotra R, Burke MF, Martyn T, Shakartzi HR, Thayer TE, ORourke C, Li P, Derwall M, Spagnolli E, Kolodziej SA, Hoeft K, Mayeur C, Jiramongkolchai P, Kumar R, Buys ES, Yu PB, Bloch KD, Bloch DB. Inhibition of bone morphogenetic protein signal transduction prevents the medial vascular calcification associated with matrix Gla protein deficiency. PLoS One 2015; 10: e0117098.
- Dan H, Simsa-Maziel S, Reich A, Sela-Donenfeld D, Monsonego-Ornan E. The role of matrix gla protein in ossification and recovery of the avian growth plate. Front Endocrinol (Lausanne) 2012; 3: 79.
- Price PA, Caputo JM, Williamson MK. Bone origin of the serum complex of calcium, phosphant, futuin, and matrix gla protein: biochemical evidence for the cancellous bone-remodeling compartment. J Bone Miner Res 2002; 17: 1171-1179.
- Atkins GJ, Welldon KJ, Wijenayaka AR, Bonewald LF, Findlay DM. Vitamain K promotes mineralization, osteoblast-to-osteocyte transition, and an anticatabolic phenotype by r-carboxylationd-dependent and independent mechanisms. Am J Physiol Cell Physiol 2009; 297: 1358-1367.