ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2012) Volume 23, Issue 3
Expression analysis of transcription factors CTCF (CCCTC- binding factor), BORIS (Brother of the Regulator Imprinted Sites) and YB-1 (Y- box binding factor 1) in osteosarcoma and glioma cell lines: Physical Interaction of CTCF~YB-1 in vivo.
Siti Zawani Mohd Ramli1*, Arunkumar Sundaram1,2*, Nurul Aini Samsuddin1*, Jafri Malin Abdullah3, Wan Faisham Nu’man Wan Ismail4, Abdul Halim Yusof4, Azmi Alias6, Mohd Saffari Mohd Haspani6, Venugopal Balakrishnan5, Nik Norliza Nik Hassan1, Shaharum Shamsuddin1, 5§
1Molecular Biology Laboratory, School of Health Sciences, Universiti Sains Malaysia, 16150 Kubang Kerian, Kelantan, Malaysia
2The Michael Smith Building, Faculty of Life Sciences, University of Manchester, Manchester M13 9PT, UK.
3Department of Neuroscience, School of Medical Sciences, Universiti Sains Malaysia, 16150 Kubang Kerian, Kelantan, Malaysia
4Department of Orthopedics, School of Medical Sciences, Universiti Sains Malaysia, 16150 Kubang Kerian, Kelantan, Malaysia
5 Institute for Research in Molecular Medicine (INFORMM), Universiti Sains Malaysia, 11800 Minden, Penang.
6Institut Kaji Saraf Tunku Abdul Rahman, Hospital Kuala Lumpur, 50586 Kuala Lumpur, Malaysia
*These authors contributed equally to this work
- Corresponding Author:
- Shaharum Shamsuddin
Institute for Research in Molecular Medicine (INFORMM)
Universiti Sains Malaysia, 11800 Minden, Penang.
Accepted date: April 19, 2016
CTCF, BORIS and YB-1 have been implicated in tumorgenesis. In this study, we examined the expressions of CTCF, BORIS and YB-1 in osteosarcoma (U2OS) and glioma (DBTRGO5MG) in comparison with their respective normal osteoblast (hFOB 1.19) and glial (SVGp12) cell lines. We examined any detected mutation for CTCF and YB1, and revealed the presence of physical in vivo CTCF~YB1 interaction. The level of CTCF mRNA was significantly higher in osteosarcoma compared to osteoblast cell line, while the level of YB-1 mRNA was similar in these cell lines. The expression of YB-1 mRNA was significantly elevated in glioma compared to glial cell line, whereas expression of CTCF mRNA was similar. The BORIS mRNA was detected only in osteosarcoma cell line. Nucleotides analysis showed undetected mutations at exon 4 and 5 (CTCF) and exon 7 (YB1) for both cancerous cell lines. Protein expression of CTCF revealed immunopositive for all cancerous and normal cell lines. YB- 1 protein expressions in glial , glioma and osteosarcoma cells was found to be immunopositive and anomalously migrated. We reported positive in vivo interaction of CTCF~YB1 protein in osteosarcoma and glioma cell lines. Different patterns of expression of CTCF,BORIS and YB- 1 mRNAs could indicate differential involvement of these transcriptional factors in tumorgenesis of osteosarcoma and glioma. Positive in vivo interaction of CTCF~YB1 in the osteosarcoma and glioma cell lines confirmed previous reports on both factors in tumor development.
Keywords
Osteosarcoma, glioma, CTCF, BORIS, YB-1, expressions
Introduction
Osteosarcoma is the most common primary malignant bone tumor in both adults and children [1]. Osteosarcoma is characterized by highly complex karyotypes and a high frequency of chromosomal copy number changes [2-4]. Over the last decades, the long-term survival of osteosarcoma patients with localized disease has improved and, with multimodal therapy, now reaches approximately 65%. However, 30-40% of patients die because of tumor progression or secondary metastases [5].
Glioma is the most common malignant brain tumor in adults [6-8]. Brain tumors arise as a result of gradual accumulation of several genetic aberrations in precursor cells which can occur either at the chromosomal level or at the gene expression level [9]. Despite considerable efforts to unravel the etiologic basis for this cancer and attempts to find a cure, glioma largely remain refractory to treatment. Except for a small percentage of cases, the tumor continue to show high morbidity and mortality [10].
Therefore, there is a need to better understand the molecular mechanisms of glioma and osteosarcoma development to improve early diagnosis, treatment and prevention of these tumours.
CTCF, YB-1 and BORIS (Brother Of the Regulator of Imprinted Sites) are all transcriptional factors that have been increasingly implicated in tumorgenesis [11, 12]. The eleven Zinc finger protein, CTCF, was found to localize at the human chromosome region, 16q22, that is well known as a cancer ‘hot spot’ [13]. CTCF is a candidate tumor suppressor gene [14, 15], and the loss of its normal function might lead to tumorgenesis [11, 16]. YB-1, a member of the YB-1 box binding protein family, is located at the human chromosome region, 1p34. YB-1 regulates both transcription and translation cascades [17] and has been demonstrated to play a critical role in various important events in carcinoma progression such as cell proliferation, metastasis, drug resistance and genotoxic stress [18-20]. Overexpression of YB-1 mRNA and protein have been reported in several malignant diseases [21-24]. BORIS, a paralogue of CTCF, is located at the human chromosome region, 20q13.2 [11]. Unlike CTCF which is ubiquitously expressed in somatic cells, the expression of BORIS is normally restricted to specific cells in testes where it may play a role in reprogramming the methylation pattern of male germ line DNA [25]. Aberrant expression of BORIS has been proposed to play a role in tumorgenesis [11] and has been reportedly detected in diverse human tumors and tumor-derived cell lines [26-29].
Although CTCF, YB-1 and BORIS have been causally linked with human malignant diseases, the involvement of these multivalent factors in osteosarcoma and glioma is still undetermined. Therefore we investigated the transcription levels of CTCF, YB-1 and BORIS mRNA and proteins to gain further insights into the role of these transcription factors in tumor osteosarcoma (U2OS) and tumor glioma (DBTRG-O5MG) cell lines.
Materials and methods
a) Cell culture and growth condition
Cell lines including tumor osteosarcoma (U2OS), tumor glioma DBTRG-O5MG (Denver Brain Tumor Research Group 05), normal osteoblast (hFOB 1.19) (Human Fetal Osteoblast) and normal glial SVG-p12 (Astrogli SV40 transformed) were purchased from ATCC, USA. U2OS cells were cultured in complete growth medium Mc Coy 5A (GIBCO@Invitrogen, USA); 10% fetal bovine serum and 1% Penicillin-Streptomycin. hFOB 1.19 cells were cultured in DMEM/F12 (GIBCO@Invitrogen, USA); 10% fetal bovine serum and 1% Penicillin- Streptomycin. DBTRG-O5MG cells were cultured in complete growth medium RPMI 1640 (GIBCO@Invitrogen, USA); 10% fetal bovine serum and 1% Penicillin- Streptomycin. SVG-p12 cells were cultured in ATCCformulated Eagle's Minimum Essential Medium (GIBCO@Invitrogen, USA); 10% fetal bovine serum and 1% Penicillin-Streptomycin. Cells were propagated in triplicate (n=3/cell line) in 75 cm2 flasks at 370C in CO2 incubator and the media were regularly changed (2-3 times a week). For RNA and protein extractions, cells were trypsinized, counted, centrifuged at 1500 rpm for 5 minutes and 107 cells were used for RNA and protein preparations.
ATTC provides consistent and low- passage cultures. They authenticates above cell lines routinely with the following test: Short tandem repeat (STR) profiling establishes a DNA fingerprint for human cell lines, monitoring the cell morphology throughout all ATCC processes, karyotyping in order to identify the species as well as variation within the cell line, isoenzyme analysis to verify the species of origin, rigorous and repeated contamination tests to ensure that the cell lines are free of mycoplasma or other bacterial or fungal agents [30].
b) Real-time PCR analysis
Total RNA from cell lines (~1x107 cells) were extracted using RNeasy Mini Kit (Qiagen, Germany) and checked for concentration, integrity and purity. Two micrograms of total RNA was reverse transcribed into cDNA using Revert Aid H Minus First Strand cDNA Synthesis Kit (Fermentas, USA). The primer sequences for CTCF, YB- 1 and GAPDH were designed using Primer 3 Input (version 0.4.0) and as follows: CTCF forward: 5’-GTC ACC CTC CTG AGG AAT CA-3’, CTCF reverse: 5’-CGT AAT CGC ACA TGG AAC AC-3’ (160 bp), YB-1 forward: 5’-GGA GAT GAG ACC CAA GGT CA-3’, YB-1 reverse: 5’-GTT AAG CCG GCA TTT ACT CA-3’ (187 bp), GAPDH forward: 5’- GAG TCA ACG GAT TTG GTC GT-3’, GAPDH reverse: 5’- TTG ATT TTG GAG GGA TCT CG-3’ (234 bp). Primers were purchased from 1st BASE, Singapore. Real-time PCR analysis with optimal condition for each gene was performed using Power SYBRGreen PCR Master Mix (ABI, USA) using Applied Biosystems 7500 Real-Time PCR System (USA).
The Real-time PCR products were also subsequently subjected to 2% agarose gel electrophoresis for the confirma tion of PCR amplicons. Serial dilution of cDNA was performed for each set of genes and primers to validate the amplification efficiencies of Real-time PCR analysis and the efficiencies were around 90-110%. Relative expression of mRNA was then determined by the comparative 2-ΔΔC T method [31]. Purified PCR products were sent for sequencing at Centre for Chemical Biology, USM, Malaysia for detection of specific mutation at particular exon.
c) SDS-PAGE and Western Blot analysis
For protein preparation, the cell pellets (~1x107 cells) were lyzed with pre-cooled radioimmunoprecipitation assay (RIPA) buffer for overnight at 4°C. The suspension was centrifuged at 12000 rpm for 20 minutes at 4°C and the supernatant was used for protein expression studies. Protein concentrations approximately 0.72 mg/mL were mixed with an equal volume of 2x loading dye/treatment buffer and heated at 950C for 5 minutes. Samples were electrophoresed in gradient (10%) SDS-PAGE and proteins were visualized via staining with commasie blue staining and western blotting. Following electrophoresis, gels were electroblotted onto PVDF membrane. The membrane was treated with 5% skim milk for 1 hour. After washing with the washing buffer, the membrane was incubated with primary antibody; anti-CTCF polyclonal, anti-YB-1 monoclonal and anti- β-actin monoclonal antibody (Santa Cruz, USA) for 1 hour at room temperature. Following three times (5 minutes each) washes in washing buffer, the membrane was treated with anti-mouse or anti-rabbit immunoglobulin antibody (Santa Cruz biotechnology) conjugated with horse radish peroxidase for 1 hour at room temperature.
A final wash was performed, and the membrane was equilibrated with detection buffer ECL solution (Milipore) for about 3 minutes and the membrane was placed in a developing folder on the hypercassette (Amersham Life Science) in a dark environment. The Kodak-X film (Sigma-Aldrich, USA) was exposed to the membrane for about 10 minutes before it was developed accordingly.
d) Co-immunoprecipitations
Protein from the total lysate was quantified and 100 μg was used for co-immuno-precipitations. The protein lysate was added with 50 μl of protein G-Sepharose (Sigma) and incubated for 1 h at 4°C. CTCF was immunoprecipitated, by adding 5 μl anti-CTCF polyclonal (Santa Cruz Biotechnology) followed by incubation for 6 hours at 4°C. The Sepharose beads were then washed five times with 1 ml of RIPA lysis buffer. Immobilized proteins were resolved by 12.5% SDS-PAGE, transferred onto PVDF membrane and probed with anti-YB-1 monoclonal antibody for western blot analysis procedure as described earlier.
Statistical analysis
Analysis was performed using the Statistical Package for Social Science, version 12.0 software. The expressions results were analyzed using Independent-T test, expressed as mean ± standard error of mean (S.E.M). A value of ‘p’ of < 0.05 was considered to be statistically significant.
Results
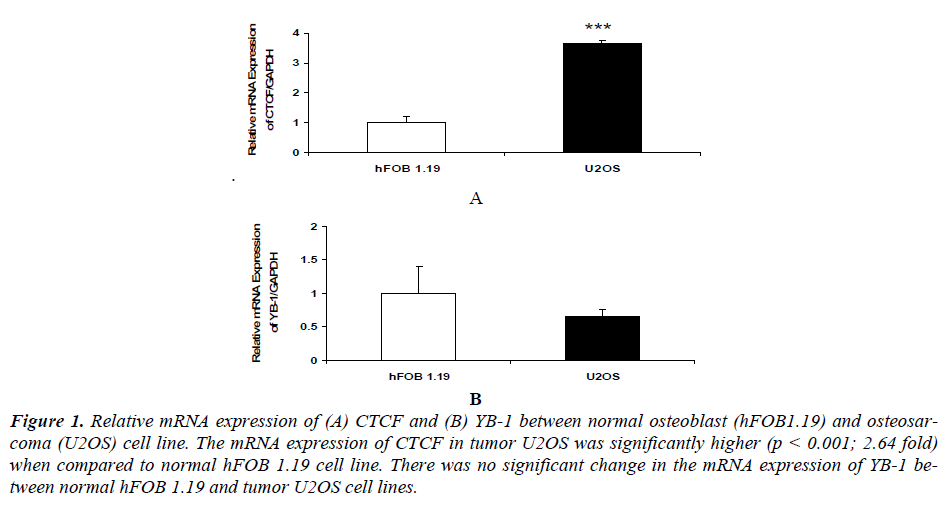
Our results indicated that mRNA expression of CTCF in osteosarcoma U2OS (p<0.01) was significantly higher compared to normal osteoblast hFOB 1.19 cell line [(Figure 1 (A)]. While there was no significant change in the mRNA expression of YB1 between normal osteoblast hFOB 1.19 and osteosarcoma U2OS cell line [Figure 1 (B)].
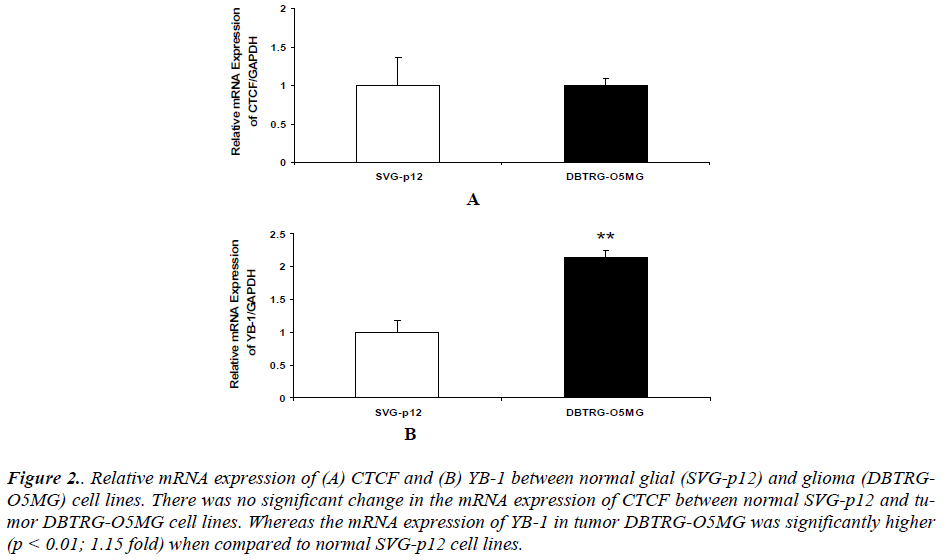
Our results also revealed that there was no significant change in the mRNA expression of CTCF between normal glial SVGp12 and glioma DBTRG-O5MG cell line (Figure 2 (A)). The mRNA expression of YB1 in glioma DBTRG-O5MG was significantly higher compared to normal glial SVGp12 cell line (Figure 2 (B)).
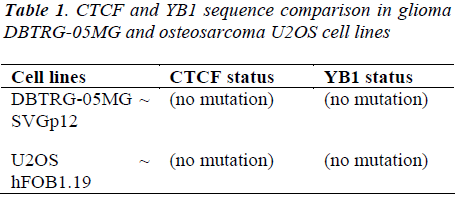
The sequence of human CTCF mRNA (160 bp) used in this study is flanking at exon 4 and exon 5 (NCBI Accession No: NM_006565.2) whereas the sequence of human YB-1 mRNA (187 bp) flanking at exon 7 (NCBI Accession No: NM_004559.3). The amplified cDNA were sent to the Sequencing facilities at the Centre for Chemical Biology USM, Penang after purification. The differences in the nucleotide sequence of CTCF and YB1 between tumor glioma DBTRG-05MG and normal glial SVGp12 cell lines and between tumor osteosarcoma U2OS and normal osteoblast hFOB 1.19 cell lines were blasted and checked for possible mutation at the respective exons. The results of CTCF sequencing between U2OS and hFOB1.19 cells [Figure 3 (A)] and between DBTRG- 05MG and SVGp12 cells [Figure 4 (A)] showed identical nucleotide sequences (99%) suggesting absence of CTCF mutation at exon 4 and 5 between these cells.
Results of YB-1 sequencing between U2OS and hFOB1.19 cells [Figure 3 (B)] and between DBTRG - 05MG and SVGp12 cells [Figure 4 (B)] showed similar nucleotide sequences (100%) suggesting the absence of mutation of YB-1 gene at exon 7 between these cells. The sequence variation was presented according to the suggestion by Human Genome Variation Society [44].
Figure 1. Relative mRNA expression of (A) CTCF and (B) YB-1 between normal osteoblast (hFOB1.19) and osteosarcoma (U2OS) cell line. The mRNA expression of CTCF in tumor U2OS was significantly higher (p < 0.001; 2.64 fold) when compared to normal hFOB 1.19 cell line. There was no significant change in the mRNA expression of YB-1 between normal hFOB 1.19 and tumor U2OS cell lines.
Figure 2. Relative mRNA expression of (A) CTCF and (B) YB-1 between normal glial (SVG-p12) and glioma (DBTRGO5MG) cell lines. There was no significant change in the mRNA expression of CTCF between normal SVG-p12 and tumor DBTRG-O5MG cell lines. Whereas the mRNA expression of YB-1 in tumor DBTRG-O5MG was significantly higher (p < 0.01; 1.15 fold) when compared to normal SVG-p12 cell lines.
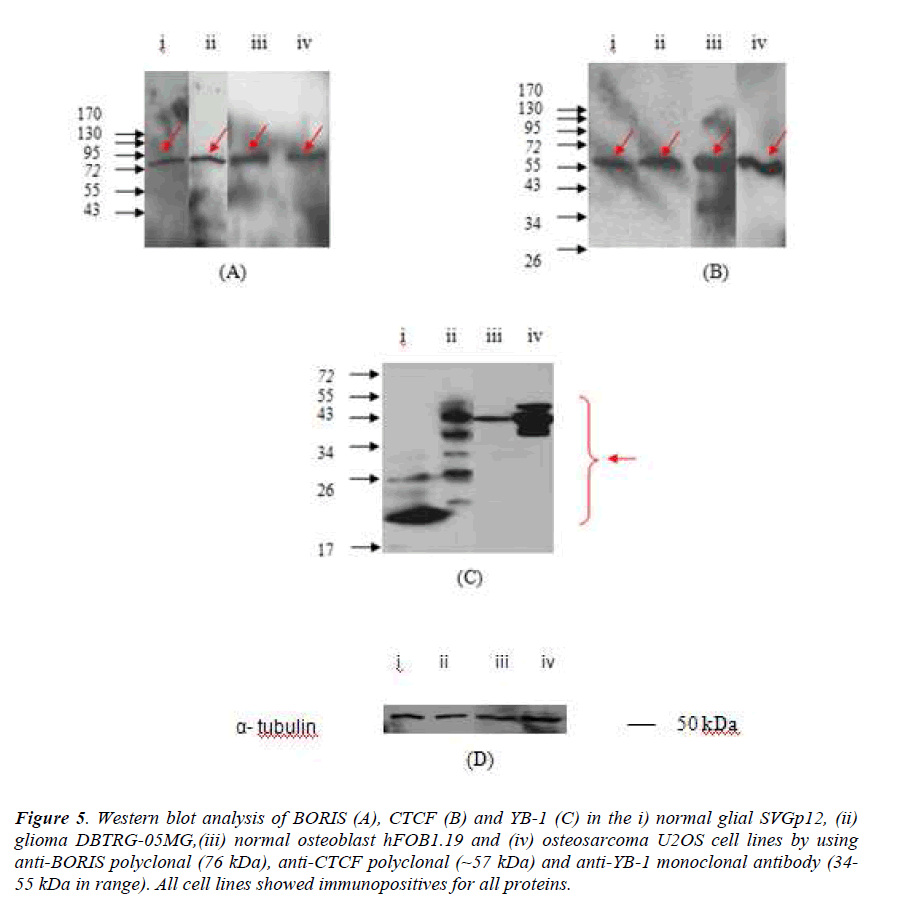
Figure 5. Western blot analysis of BORIS (A), CTCF (B) and YB-1 (C) in the i) normal glial SVGp12, (ii) glioma DBTRG-05MG,(iii) normal osteoblast hFOB1.19 and (iv) osteosarcoma U2OS cell lines by using anti-BORIS polyclonal (76 kDa), anti-CTCF polyclonal (~57 kDa) and anti-YB-1 monoclonal antibody (34- 55 kDa in range). All cell lines showed immunopositives for all proteins.
Western Blotting of SVGp12, DBTRG-05MG, hFOB1.19 and U2OS cell lines (Figure 5) showed immunopositive for CTCF, YB-1 and BORIS proteins. Total lysates were separated in 12.5 % SDS-PAGE, then probed with α- BORIS polyclonal antibody, α-CTCF polyclonal antibody and α-YB-1 monoclonal antibody. The presence of BORIS protein is indicated by a red arrow at ~76 kDa for all cell lines. The presence of CTCF protein is indicated by a red arrow at ~57 kDa for all cell lines and the presence of YB-1 is indicated by a red arrow at ~17 kDa to ~ 26 kDa for SVGp12; ~17 kDa to ~55 kDa for DBTRG-05MG; ~43 kDa for hFOB1.19 and ~43 kDa to ~72 kDa for U2OS total lysate.
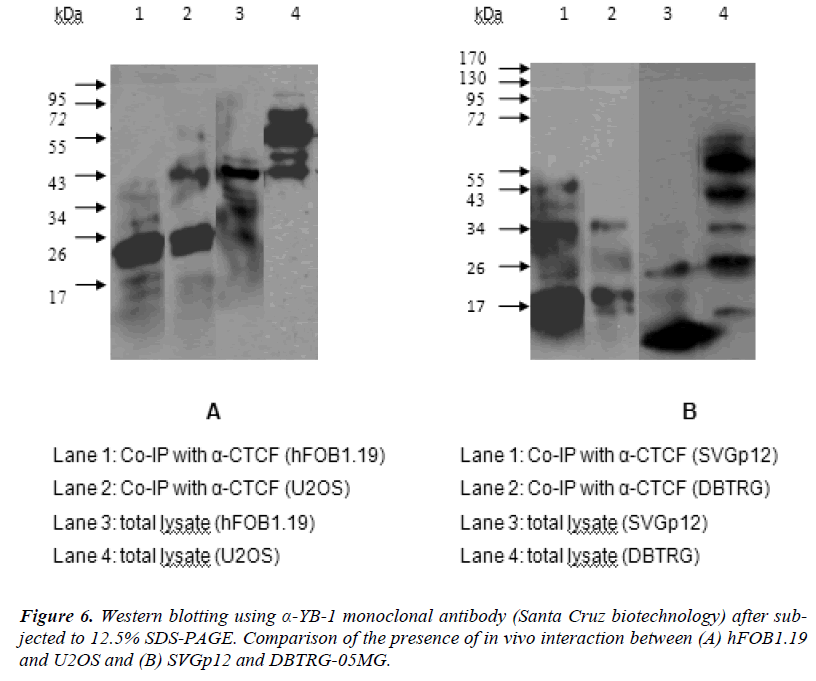
In order to evaluate the interaction of CTCF and YB1 at in vivo condition, co- immunoprecipitation assays have been carried out with the extracts from hFOB1.19, U2OS, SVGp12 and DBTRG-05MG cells. Proteins bound by anti-CTCF were incubated with protein G-Sepharose- 4BFast, washed, separated on SDS-PAGE, and analyzed by ECL immunoblotting. Fig. 6 (A) (lane 1, 2) shows that CTCF immunoprecipitates obtained from hFOB1.19 (lane 1) or U2OS (lane 2) contain the YB-1 bands co-migrating with the YB-1 protein from the whole hFOB1.19 cell lysate (lane 3) and whole U2OS cell lysate (lane 4). Figure 6 (B) (lane 1, 2) also shows that CTCF immunoprecipitates obtained from SVGp12 (lane 1) or DBTRG-05MG (lane 2) contain the YB-1 bands co-migrating with the YB-1 protein from the whole SVGp12 cell lysate (lane 3) and whole DBTRG-05MG cell lysate (lane 4).
Discussion
The significantly increased mRNA expression of CTCF might promote cell survival by protecting osteosarcoma tumor cells from apoptosis, whilst suppression of apoptosis is a critical factor supporting tumor progression [32, 33]. Similar results of elevated levels of CTCF in breast cancer cells and tumors have been reported and the heightened levels were associated with resistance to apoptosis [34]. Over-expression of YB-1 mRNA has been reported earlier in various types of cancers such as pancreatic adenocarcinoma, metastatic prostate carcinoma, ovarian carcinoma, medulloblastoma [24] and pediatric glioblastoma [35]. YB-1 regulates both transcription and translation cascades [17] which is thought to play a critical role in various important events in carcinoma progression [18- 20]. Therefore, the augmented expressions of YB-1 mRNA in DBTRG-O5MG cell line could suggest a crucial link of YB-1 in tumor glioma. We showed BORIS mRNA was able to be expressed in osteosarcoma U2OS cells and was not found to be detected in normal osteoblast hFOB1.19 cell line [36]. This expression data is consistent with other evidence in the literature that BORIS is a true cancer/ testis gene in which expression occurs only in malignancies and testis, and not in other tissues [25]. Previous studies have shown aberrant expression of BORIS mRNA in various human cancers such as breast, melanoma, neuroblastoma, prostate and colon cell lines [27], breast, prostate and colon tumors [27], osteosarcoma tumors [26], uterine cancers [29] and leukocytes of breast cancer patients [28]. The appearance of common cancer testis antigen (including BORIS) during gametogenesis and tumorigenesis prompts the hypothesis that induction of the gametogenetic programme in somatic cells may be associated with tumour development [37, 38]. Thus, our results postulate that aberrant mRNA expression of BORIS in osteosarcoma cell line could imply its potential role in this tumor. On the other hand, the expression of BORIS mRNA was not found in glioma DBTRG-05MG and normal glial SVGp12 cell lines [36]. Recently Hines et al., 2010 [39] has reported that BORIS mRNA was not expressed in most human breast cancer cell lines and tumors. Furthermore expression of BORIS mRNA showed no significant difference between normal and prostate cancer tissues overall and no relationship was seen to clinical parameters [40]. Therefore, until this stage we reported the undetected expression of BORIS in glioma cell line [36].
The sequencing results of cDNAs obtained from CTCF flanking at exon 4 and 5, and YB-1 flanking at exon 7 were found to be identical between the glioma DBTRG- 05MG and glial cell lines SVGp12 and between the osteosarcoma U2OS and osteoblast cell lines hFOB1.19 thus confirmed that there are no sequence changing in the respective particular exons for both markers (Table 1).
Proteomics study showed YB1 protein migrated anomalously for glial SVGp12, glioma DBTRG-05MG and osteosarcoma U2OS cell lines. Thus, suggesting post translational modification of the YB1 protein with high expression in the three cell lines. For comparison, we can see the high expression of YB1 mRNA and YB1 protein in glioma DBTRG- 05MG compared to the glial SVGp12 cell lines. Interestingly the high expression of YB1 protein for osteosarcoma U2OS unparallel with the result of YB1 mRNA expression for the cell line. Again, this might be due to the post translational modification of the YB1 protein in the osteosarcoma U2OS cell lines
The Co-IP results showed the presence of extra bands (sub units) at the range of 19kDa to 72kDa and the proteins were actually the targeted YB-1 protein. It was assumed to be the targeted protein because the detection of YB-1 was done using mouse monoclonal antibody. The apparent molecular weights of the YB-1 (range 19kDa to 72kDa) are wide in range which migrated anomalously; in comparison to the published size (50kDa) (Santa Cruz biotechnology) and previously reported (50kDa) [41]. These additional lower and higher mobility bands/ sub units represent a post translationally modified form of YB-1 that is most apparent when YB-1 protein is highly over-expressed [42]. These suggest YB-1 protein binds to CTCF protein in all cell lines and this interaction may occur direct or indirect.
As a conclusion, elevated levels of CTCF together with expressed BORIS mRNA in U2OS cell line [36] and higher YB-1 mRNA in DBTRG-05MG cell line could indicate its potential role in tumorgenesis. YB-1 mRNA expression in U2OS cell line and CTCF mRNA expression in DBTRG-05MG cell line was similar compared to their normal counterparts suggesting that more studies are required to understand their possible functional link in these tumors. Studying the expression of CTCF/ BORIS and YB-1 in glioma DBTRG-05MG/ U2OS osteosarcoma cell lines, we encountered the undetected mRNA expression of BORIS in glioma cell line although the protein expression of the BORIS in the glioma cell line is positive. Thus, we should focus deeper on molecular mechanism/ pathway on BORIS expression with consideration of its mRNA characteristic, with involvement of complex mechanism, BORIS as the cell- dependent gene, its alternative splice variants and promoter- dependent characteristics [45]. In addition, undetected mRNA BORIS in breast tumors cells has been reported by Hines et al. (2010) [39] and this finding is relevant to our undetected mRNA BORIS in the glioma DBTRG-05MG cell line. Previously, a study by our co-author has reported biological relevance of CTCF/ YB-1 interactions, indicating that deregulation or their abnormal interactions could play an important role in tumor development [43]. Therefore, evaluation of CTCF/ YB1 interactions in U2OS and DBTRG-05MG cancer cell lines will further ascertain its potential functional roles in these cancers.
Acknowledgements
We thank all the students and staffs of Molecular Biology Laboratory, School of Health Sciences, Department of Neurosciences, School Of Medical Sciences / Brain Mind Nexus., Universiti Sains Malaysia Health Campus. We are grateful to the technical staffs of the Craniofacial laboratory, School of Dental Science, USM Health Campus for their assistance with the inverted microscopy. We thank the Institute for Molecular Medicine (INFORMM), USM Health Campus for allowing us to use their tissue culture facilities. Special thanks to all the neurosurgeons, nurses and technical staffs in operation theater 15 (Neurosurgery), Hospital Universiti Sains Malaysia (HUSM).
Grant Support. This study was supported by eScience-Fund (305/PPSK/6113213), Ministry of Science & Innovation (MOSTI) Malaysia, Fundamental Research Grant Scheme (FRGS) grant (2003/PPSP/6171108) and the Ministry of Science & Innovation, Malaysia for the National Science Fellowship (to support the postgraduate study of S.Z.M.R)
References
- Bell W, Siegal G: Osteosarcoma. In: Cullinane C, Burchill S, Squire J, Lewis I, O'Leary J, editors. Molecular biology and pathology of paediatric cancer. London: Oxford University Press.; 2002.
- Bayani J, Zielenska M, Pandita A, Al-Romaih K, Karaskova J, Harrison K, Bridge JA, Sorensen P, Thorner P, Squire JA: Spectral karyotyping identifies recurrent complex rearrangements of chromosomes 8, 17, and 20 in osteosarcomas. Genes Chromosomes Cancer 2003; 36: 7-16.
- Man TK, Lu XY, Jaeweon K, Perlaky L, Harris CP, Shah S, Ladanyi M, Gorlick R, Lau CC, Rao PH: Genome-wide array comparative genomic hybridization analysis reveals distinct amplifications in osteosarcoma. BMC Cancer 2004; 4: 45.
- Man TK, Chintagumpala M, Visvanathan J, Shen J, Perlaky L, Hicks J, Johnson M, Davino N, Murray J, Helman L, et al: Expression profiles of osteosarcoma that can predict response to chemotherapy. Cancer Res 2005; 65: 8142-8150.
- Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M, Winkelmann W, et al: Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol 2002; 20: 76-790.
- Machado CM, Schenka A, Vassallo J, Tamashiro WM, Goncalves EM, Genari SC, Verinaud L: Morphological characterization of a human glioma cell l ine. Cancer Cell Int 2005; 5: 13.
- Efird JT: Season of birth and risk for adult onset glioma. Int J Environ Res Public Health 2010; 7: 1913- 1936.
- Jansen M, Yip S, Louis DN: Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol 2010; 9: 717-726.
- Furnari FB, Huang HJ, Cavenee WK: Genetics and malignant progression of human brain tumours. Cancer Surv 1995; 25: 233-275.
- Wrensch M, Fisher JL, Schwartzbaum JA, Bondy M, Berger M, Aldape KD: The molecular epidemiology of gliomas in adults. Neurosurg Focus 2005; 19: E5.
- Klenova EM, Morse HC, 3rd, Ohlsson R, Lobanenkov VV: The novel BORIS+CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol 2002; 12:399-414.
- Kuwano M, Uchiumi T, Hayakawa H, Ono M, Wada M, Izumi H, Kohno K: The basic and clinical implications of ABC transporters, Y-box-binding protein-1 (YB-1) and angiogenesis-related factors in human malignancies. Cancer Sci 2003; 94: 9-14.
- Filippova GN, Lindblom A, Meincke LJ, Klenova EM, Neiman PE, Collins SJ, Doggett NA, Lobanenkov VV: A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes Chromosomes Cancer 1998; 22: 26-36.
- Rasko JE, Klenova EM, Leon J, Filippova GN, Loukinov DI, Vatolin S, Robinson AF, Hu YJ, Ulmer J, Ward MD, et al: Cell growth inhibition by the multifunctional multivalent zinc-finger factor CTCF. Cancer Res 2001; 61: 6002-6007.
- Qi CF, Martensson A, Mattioli M, Dalla-Favera R, Lobanenkov VV, Morse HC, 3rd: CTCF functions as a critical regulator of cell-cycle arrest and death after ligation of the B cell receptor on immature B cells. Proc Natl Acad Sci U S A 2003; 100: 633-638.
- Ohlsson R, Renkawitz R, Lobanenkov V: CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet 2001; 17: 520- 527.
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M: The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays 2003; 25: 691-698.
- Ise T, Nagatani G, Imamura T, Kato K, Takano H, Nomoto M, Izumi H, Ohmori H, Okamoto T, Ohga T, et al: Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res 1999; 59: 342-346.
- Shibao K, Takano H, Nakayama Y, Okazaki K, Nagata N, Izumi H, Uchiumi T, Kuwano M, Kohno K, Itoh H: Enhanced coexpression of YB-1 and DNA topoisomerase II alpha genes in human colorectal carcinomas. Int J Cancer 1999; 83: 732-737.
- Levenson VV, Davidovich IA, Roninson IB: Pleiotropic resistance to DNA-interactive drugs is associated with increased expression of genes involved in DNA replication, repair, and stress response. Cancer Res 2000; 60: 5027-5030.
- Janz M, Harbeck N, Dettmar P, Berger U, Schmidt A, Jurchott K, Schmitt M, Royer HD: Y-box factor YB-1 predicts drug resistance and patient outcome in breast cancer independent of clinically relevant tumor biologic factors HER2, uPA and PAI-1. Int J Cancer 2002; 97: 278-282.
- Ito Y, Yoshida H, Shibahara K, Uruno T, Nakano K, Takamura Y, Miya A, Kobayashi K, Yokozawa T, Matsuzuka F, et al: Y-box binding protein expression in thyroid neoplasms: its linkage with anaplastic transformation. Pathol Int 2003; 53: 429-433.
- Gimenez-Bonafe P, Fedoruk MN, Whitmore TG, Akbari M, Ralph JL, Ettinger S, Gleave ME, Nelson CC: YB-1 is upregulated during prostate cancer tumor progression and increases P-glycoprotein activity. Prostate 2004; 59:337-349.
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM: ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004; 6: 1-6.
- Loukinov DI, Pugacheva E, Vatolin S, Pack SD, Moon H, Chernukhin I, Mannan P, Larsson E, Kanduri C, Vostrov AA, et al: BORIS, a novel male germ-linespecific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci USA 2002; 99: 6806-6811.
- Ulaner GA, Vu TH, Li T, Hu JF, Yao XM, Yang Y, Gorlick R, Meyers P, Healey J, Ladanyi M, Hoffman AR: Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methyla tion changes of a CTCF-binding site. Hum Mol Genet 2003; 12: 535-549.
- Vatolin S, Abdullaev Z, Pack SD, Flanagan PT, Custer M, Loukinov DI, Pugacheva E, Hong JA, Morse H, 3rd, Schrump DS, et al: Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res 2005; 65: 7751-7762.
- D'Arcy V, Abdullaev ZK, Pore N, Docquier F, Torrano V, Chernukhin I, Smart M, Farrar D, Metodiev M, Fernandez N, et al: The potential of BORIS detected in the leukocytes of breast cancer patients as an early marker of tumorigenesis. Clin Cancer Res 2006; 12: 5978-5986.
- Risinger JI, Chandramouli GV, Maxwell GL, Custer M, Pack S, Loukinov D, Aprelikova O, Litzi T, Schrump DS, Murphy SK, et al: Global expression analysis of cancer/testis genes in uterine cancers reveals a high incidence of BORIS expression. Clin Cancer Res 2007; 13: 1713-1719.
- ATCC Cultures: Maintaining high standards in Cell Culture, www.atcc.org
- Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402-408.
- Evan GI, Vousden KH: Proliferation, cell cycle and apoptosis in cancer. Nature 2001; 411: 342-348.
- Igney FH, Krammer PH: Death and anti-death: tumour resistance to apoptosis. Nat Rev Cancer 2002; 2: 277- 288.
- Docquier F, Farrar D, D'Arcy V, Chernukhin I, Robinson AF, Loukinov D, Vatolin S, Pack S, Mackay A, Harris RA, et al: Heightened expression of CTCF in breast cancer cells is associated with resistance to apoptosis. Cancer Res 2005; 65: 5112-5122.
- Faury D, Nantel A, Dunn SE, Guiot MC, Haque T, Hauser P, Garami M, Bognar L, Hanzely Z, Liberski PP, et al: Molecular profiling identifies prognostic subgroups of pediatric glioblastoma and shows increased YB-1 expression in tumors. J Clin Oncol 2007; 25: 1196-1208.
- Ramli SZM, Shamsuddin S, Hassan NN, Alias A, Haspani MSM and Abdullah J (2011). The Expression of BORIS Protein in a Newly Established Primary Glioma Cell Culture Line, Molecular Targets of CNS Tumors, Miklos Garami (Ed.), ISBN: 978-953-307- 736-9, InTech, Available from: http://www.intechopen.-com/articlesshow/ title/the-expression-of-boris-protein-in-a-newlyestablished- primary-glioma-cell-culture-line
- Old LJ. Cancer/testis (CT) antigens - a new link between gametogenesis and cancer. Cancer Immun 2001; 1: 1.
- Kalejs M, Erenpreisa J: Cancer/testis antigens and gametogenesis: a review and "brain-storming" session. Cancer Cell Int 2005; 5: 4.
- Hines WC, Bazarov AV, Mukhopadhyay R, Yaswen P: BORIS (CTCFL) is not expressed in most human breast cell lines and high grade breast carcinomas. PLoS One 2010; 5:e9738.
- Hoffmann MJ, Muller M, Engers R, Schulz WA: Epigenetic control of CTCFL/BORIS and OCT4 expression in urogenital malignancies. Biochem Pharmacol 2006; 72: 1577-1588.
- Gessner C, Woischwill C, Schumacher A, Liebers U, Kuhn H, Stiehl P, Jürchott K, Royer HD, Witt C, Wolff G: Nuclear YB-1 expression as a negative prognostic marker in nonsmall cell lung cancer. European Respiratory Journal 2004; 23: 14-19.
- Al-Kandari W, Koneni R, Navalgund V, Aleksandrova A, Jambunathan S, Fontes JD: The Zinc Finger Proteins ZXDA and ZXDC Form a Complex that Binds CIITA and Regulates MHC II Gene Transcription. Journal of Molecular Biology 2007; 369: 1175-1187.
- Chernukhin IV, Shamsuddin S, Robinson AF, Carne AF, Paul A, El-Kady AI, Lobanenkov VV, Klenova EM: Physical and Functional Interaction between Two Pluripotent Proteins, the Y-box DNA/RNA-binding Factor, YB-1, and the Multivalent Zinc Finger Factor, CTCF. The Journal of Biological Chemistry 2000; 275: 29915-29921.
- den Dunnen JT and Antonarakis SE. Mutation Nomenclature Extensions and Suggestions to Describe Complex Mutations: A Discussion. Human Mutation 2000; 5: 7 - 12
- Renaud S, Pugacheva EM, Delgado MD, Braunschweig R, Abdullaev Z, Loukinov D, Benhattar J, Lobanenkov V. Expression of the CTCF-paralogous cancer- testis gene, brother of the regulator of imprinted sites (BORIS), is regulated by three alternative promoters modulated by CpG methylation and by CTCF and p53 transcription factors. Nucleic Acids Res 2007; , 35 (21): 7372-7388