ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 15
Evolutionary analysis of the antigenic determinant, glycosylation, and sialidase sites of neuraminidase from the human influenza A virus isolated in China from 1995 to 2012
Hongmei Zhang1, Hao Li1, Lei Zhao1, Xiaoyu Zhang2, Jianfang Wang1, Shelan Liu3 and Lili Chu4*
1Clinical Research Center, the Second Affiliated Hospital of Southeast University, Nanjing, Jiangsu, PR China
2Department of Hepatology, the Second Affiliated Hospital of Southeast University, Nanjing, Jiangsu, PR China
3Department of Infectious Diseases, Zhejiang Center for Disease Control and Prevention, Hangzhou, Zhejiang, PR China
4Pediatric Research Institute, Children’s Hospital of Nanjing Medical University, Nanjing, Jiangsu, PR China
- *Corresponding Author:
- Lili Chu
Pediatric Research Institute
Nanjing Medical University Affiliated Nanjing Children’s, Hospital, PR China
Accepted on June 2, 2017
Background: Improve the understanding of evaluative characteristics of the NA gene, NA protease active sites, and NA glycosylation sites in H1N1 virus isolated from China during 1995-2012.
Method: A total of 200 NA gene sequences were downloaded from the National Center for Biotechnology Information’s GenBank. Phylogenetic analyses, multiple sequence alignment, glyco-sylation sites analyses, and protein structure prediction were conducted with Mega5.05, ClustalW, NetNGlyc1.0 program, and SWISS-MODEL server, respectively.
Results: The NA genes of the Chinese pandemic H1N1 2009 virus were different from those of 1995-2008 seasonal sequences (low similarity of 75.5%-77.1%), but were similar to strains from other countries (high similarity of 99.1%-99.9%). The Chinese resistant strains were close to the wild-type strains but had low similarity from other countries. Six glycosylation sites (44 (NHT), 50 (NQS), 58 (NST), 63 (NHT), 70 (NNT), 434 (NTT), and 455 (NWS)) significantly changed, and four other sites remained stable. A total of 8 and 11 NA sialidase activity and auxiliary sites were conserved, respectively, except for the H275Y substitutions in 11 strains. The most common mutations were R222Q, V234M, D344N, and K254R. H275Y, despite its missing glycosylation sites, can change its 3D structure.
Conclusions: Except for sialidase, some of the antigenic determinants and glycosylation sites from Chinese H1N1 influenza NA genes have changed in the past 20 y, which is related to the periodic outbreak in China.
Keywords
H1N1 influenza, Phylogenic characteristics, Neuraminidase gene.
Introduction
Influenza A is one of the most common respiratory diseases. It has caused four pandemics in the past century; occurred in 1918 (Spanish, H1N1), 1957 (Asia, H2N2), 1968 (Hong Kong, H3N2), and 2009 (H1N1). These pandemics resulted in millions of deaths and considerable economic losses. The 2009 influenza outbreak, which was caused by a new strain of human influenza A (H1N1), was the first influenza pandemic in the 21st century. This novel virus spread worldwide and caused approximately 17,000 deaths at the beginning of 2010 [1-4]. This virus is no longer considered “novel,” but it is still circulating in many countries. An important reason for the periodic breakout is the variation in the virus structure and important sites.
On the surface of the H1N1 virus, there are two important antigens: hemagglutinin and Neuraminidase (NA). NA combines with sialic acid on the surface of infected cells, thus promotes the release of the virus from the infected cell membrane and prevents virus aggregation. These processes allow the virus to spread and enhance the infection. The active site on NA has highly conserved amino acids that are the key targets of anti-influenza drugs, such as oseltamivir (Tamiflu) [5,6]. Tamiflu-resistant cases have been reported in Canada, Denmark, Japan, and China during 2009-2012 [7]. These reports indicate that the NA gene of the H1N1 virus has mutated. Asia, particularly China, is believed to be the source of pandemic influenza. Investigating the evolutionary characteristics and profiles of Chinese influenza A has been highly important. In this study, we analyse the evolution of the NA gene, NA protease active sites, and NA glycosylation sites in the H1N1 virus to reveal its future evolutionary trends and provide novel insights into NA critical site-based drug design for the development of novel therapeutics.
Materials and Methods
NA gene sequence sources
A total of 200 NA gene and protein sequences of human H1N1 strains from 1995-2012 were randomly downloaded from National Center for Biotechnology Information’s (NCBI) Influenza Virus Resource (http://www.ncbi.nlm.nih.gov/ genomes/FLU/FLU.html). Fifty NA sequences with H275Y substitutions, of which 11 are identical to the 200 sequences, and were also randomly selected from the Virus Resource. In total, 114 and 125 strains isolated from China and other countries (including two vaccine strains recommended by the World Health Organization, namely, A/California/07/2009 and A/Brisbane/59/2007) were analysed respectively in this study.
NA gene analysis
Phylogenetic analyses were conducted with Mega5.05. Evolutionary history was inferred through the neighbor joining method. An optimal tree with the sum of branch lengths equal to 0.44414677 was created. The tree was drawn with scales and branch lengths in the same units as those of the evolutionary distances were utilized to construct the phylogenetic tree. Evolutionary distances were computed with the maximum composite likelihood method and were expressed in the units of the number of base substitutions per site. The first, second, third, and noncoding codon positions were all included. All positions containing gaps and missing data were eliminated from the dataset (i.e., complete deletion option). A total of 1406 positions were included in the final dataset.
NA genes were analysed with the software ClustalW (http:// www.ebi.ac.uk/Tools/clustalw2/index.html). Analysis of the Nglycosylation sites was conducted with the NetNGlyc1.0 program (http://www.cbs.dtu.dk/ services/NetNGlyc/).
Prediction of the 3D structure of the NA protein
To investigate whether glycosylation site composition influences the 3D structure of the NA protein, we used two representative strains, namely, Beijing-2009 (A/Beijing/ 3/2009, GQ225383) and Taiwan-1999 (A/Taiwan/5072/1999, CY040140), to construct a 3D model with the software SWISS-MODEL [8] (available through the web interface at http://swissmodel.expasy.org/). Visual diagrams were constructed with the program Visual Molecular Dynamics version 1.8.6, which is available for free at http:// www.ks.uiuc.edu/Research/vmd/ [9].
We also investigated whether the H275Y drug-resistant site influences the NA protein structure. We selected one wild strain, namely, Beijing-2009 (A/Beijing/3/2009, GQ225383), and one resistant seasonal strain, namely, Gansu Chenguan-2007 (A/Gansu Chenguan/1129/2007, EU879064),to construct the spiral structure. Analyses were performed as described above.
Results
Comparison of H1N1 NA sequences isolated from China and other countries
The sequences of the NA gene of A/H1N1 influenza virus strains were downloaded from the NCBI database. A total of 100 H1N1 strains from different provinces/cities of China and 100 H1N1 strains isolated from other countries during 1995-2012 were analysed. The phylogenetic trees can be divided into two distinct branches. One branch consists of the 1995-2008 strains with A/Brisbane/59/2007; it is elongated upward and clustered by the year of isolation, not by area or country. This branch shows the strains clustered chronologically. All of the 2009-2012 novel H1N1 from different countries are clustered in another branch with A/ California/07/2009, which varies considerably from the past influenza NA genes that circulated in the population, except for four strains in 2009 (A/New York/3442/2009, A/Hong Kong/ 15273/2009, A/Hong Kong/25119/2009, and A/Hong Kong/ 46035/2009). Interestingly, compared with A/California/ 07/2009 at the AA level, all of the four strains showed H275Y mutation.
The NA genes of 2009-2011 Chinese H1N1 strains were 99.1%-99.9% homologous to those of strains isolated from other countries or regions, such as Thailand, Karaj, Athens, Nether-lands, Mexico City, California, Houston, New York, and Boston during 2009-2012. However, the novel Chinese strains exhibited only 75.5%-77.1% similarity to the strains collected from 1995-2008 in China.
Molecular evolution of the H1N1 NA gene isolated from humans in China from 1995–2012
NA antigenic determinant substitutions: The 200 human NA amino acid sequences that were isolated from China and other countries (100 sequences each) were analysed with the DNASTAR software. The most common anti-gen substitutions in the NA gene observed in the sequences from China during the past 18 y were M23I (38%, 76/200), H45Q (73%, 36.5/200), D79S (36.5%, 73/200), T81V (36.5%, 73/200), T84K (36.5%, 73/200), V149I (36.5%, 73/200), R222N (37%, 74/200), N248D (38.5%, 77/200), V321I (41.5%, 83/200), D344N (40.5%, 81/200), L430R (39.5%, 79/200), R432K (39.5%, 79/200), T436X (36.5%, 73/200), A454V (35.5%, 71/200), and N455G (36%, 72/200). Most of the other Antigenic Determinant (AD) sites were conserved in different years.
NA sialidase activity site substitutions: The center of NA sialidase activity includes eight catalytic sites that are conserved in all NA subtypes. These sites are R118, D151, R152, R225, E277, R293, R368, and Y402 (in N1 numbering); they directly interact with the substrate and framework sites (E119, R156, W179, S180, D/N199, I223, E228, H275, E278,N295, and E425) that support catalytic residues [10-12]. No substitutions that may have affected the NA sialidase activity were observed, except in 11 strains (A/Gansu/Chenguan/ 1129/2007, A/Hong Kong/FFD/2009, A/Hong Kong/ 15273/2009, A/Hong Kong/25119/2009, A/Hong Kong/ 46035/2009, A/Taiwan/90252/2011, A/Belgium/G257/08, A/ Managua/954.02/2008, A/Managua/3153.01/2008, A/Parma/ 34/08, and A/New York/3442/2009) that exhibited H275Y mutation (11/200, 5.5%) and 1 strain (A/TW/130/96) that exhibited E119K mutation (1/200, 0.5%). H275Y mutation is resistant to Tamiflu. The other amino acids were conserved in all 19 sites that are important for NA activity.
NA N-glycosylation site substitutions: We obtained series scores for the potential glycosites by submitting the alignment files to a prediction server. In consideration of experimental errors, occasional glycosites that may result from the genomic sequencing or translated by different genetic codes were excluded [13].
The NA protein has 10 main potential glycosylation sites, namely, amino acids 44 (NHT), 50 (NQS), 58 (NST/NNT), 63 (NHT/NQT), 70 (NNT), 88 (NSS), 146 (NGT), 235 (NGS), 434 (NTT), and 455 (NWS) [10,14]. All influenza A H1N1 strains that were new in 2009 had the same distribution of glycosylation sites. Two (2/100, 2%) Chinese strains and 69 (69/100, 69%) strains from other countries showed glycosylation site 44 (NHT) and were all isolated before 2008, except for one 2009 strain. Interestingly, two and four additional N-linked glycosylation sites in the NA stalk region (42 (NQS)) were found in Chinese and foreign (2% and 4%, respectively) NA sequences because of N44S mutation after 2010 (A/Beijing/3907/2010, A/Taiwan/611/2011, A/ Thailand/Cu-H2543/2010, A/Bangkok/INS520/2010, A/ Boston/DOA93/2012, and A/Karaj/5718/2010). Glycosylation sites 50 (NQS) and 63 (NQT) increased, whereas 63 (NHT), 70 (NNT), 434 (NTT), and 455 (NWS) decreased gradually. Glycosylation sites 44 (NHT), 70 (NNT), 434 (NTT), and 455 (NWS) were not present in many of the 2009 strains, but distinct changes were observed in 50 (NQS), 58 (NNT), and 63 (NQT). The three other sites (88 (NSS), 146 (NGT), and 235 (NGS)) were conservative for all NAs (Table 1).
| Country | Time periods (numbers) | 44 NHT | 50 NQS | 58 NST/NNT | 63 NHT/NQT | 70 NNT | 88 NSS | 146 NGT | 235 NGS | 434 NTT | 455 NWS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| China | 1995-2008 (24) | 2 (3.57) | 0 (0.00) | 23/1 (95.83)/(4.17) | 20/0 (83.33)/(0.00) | 2 (8.34) | 24 (100) | 24 (100) | 24 (100) | 23 (95.83) | 23 (95.83) |
| 2009 (56) | 0 (0.00) | 8 (14.29) | 3/53 (5.37)/(94.64) | 10/8 (17.86)/(14.29) | 3 (5.37) | 56 (100) | 56 (100) | 56 (100) | 2 (3.57) | 3 (5.37) | |
| 2010-2012 (20) | 0 (0.00) | 6 (30) | 3/53 (5.37)/(94.64) | 0/6 (0.00)/(30.00) | 0 (0.00) | 20 (100) | 20 (100) | 20 (100) | 0 (0.00) | 0 (0.00) | |
| 1995-2008 (84) | 68 (80.95) | 0 (0.00) | 76/1 (90.48)/(1.19) | 68/14 (80.95)/(16.67) | 84 (100) | 84 (100) | 84 (100) | 84 (100) | 73 (86.90) | 83 (98.80) | |
| Other countries | 2009 (7) | 1 (14.29) | 5 (71.43) | 2/5 (28.57)/(71.43) | 3/4 (42.86)/(57.14) | 2 (28.57) | 7 (100) | 7 (100) | 7 (100) | 4 (57.14) | 2 (28.57) |
| 2010-2012 (9) | 0 (0.00) | 8 (88.89) | 0/7 (0.00) | 3/4 (42.86)/(57.14) | 3 (33.33) | 9 (100) | 9 (100) | 9 (100) | 0 (0.00) | 2 (22.22) |
Table 1. Distribution of the potential glycosylation sites in neuraminidase that was isolated from China and other countries during 1995-2012.
Comparison of NA-resistant strains from China and other countries
To compare the NA-resistant strains (H275Y) from China and other countries and to examine if certain characteristic evolutionary changes occurred, phylogenetic analyses of the NA gene of the 20 NA-resistant strains isolated from China, 30 NA-resistant strains isolated from other countries, and 3 wildtype strains (275 H) were performed with Mega5.05. The phylogenetic tree of the 3 wild-type strains and 50 NAresistant strains can be divided into two main branches. The 20 NA-resistant strains and 3 wild-type strains isolated from China clustered in one branch. The remaining 30 H275Y resistant strains from other countries clustered closely in another branch. Furthermore, different sub-species were observed even though the strains were isolated from the same area. With the strains isolated from Hong Kong in 2009 as an ex-ample, several strains clustered with the strains from Gansu or Taiwan, whereas several other strains clustered with the strains from Hong Kong in 2008 (Figure 1).
Prediction of the 3D structure of H1N1 NA in wildtype and resistant strains
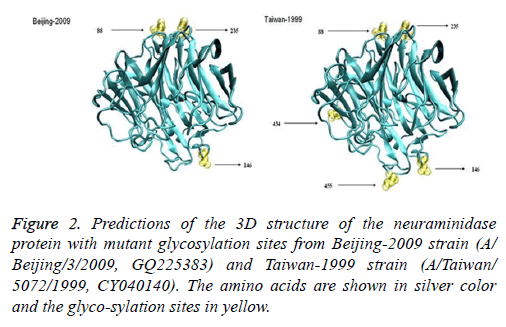
Six protein sequences were analysed; the sequences included one strain with the glycosylation sites deleted (A/Beijing/ 3/2009 (H1N1)), one strain that was not lacking any glycosylation sites (A/Taiwan/5072/1999 (H1N1)), two H275Y resistant strains (A/Hong Kong/17566/2009 (H1N1) and A/ Gansu/Chenguan/1129/2007 (H1N1)), and two wild-type strains (A/Beijing/3/2009 (H1N1) and A/Nanchang/4/1998 (H1N1)). We compared the 3D structure of the wild-type and mutant strains isolated from humans. Both strains should be composed of silk-like random coils, helices, a folded sheet, and a rolled angle. This composition indicates that glycosylation sites 88, 146, 235, and 455 are located on the surface of the protein, but glycosylation site 434 is located inside the structure. A/Beijing/3/2009 (H1N1) had three glycosylation sites that were identical to those of the A/Taiwan/5072/1999 (H1N1) strain. However, this strain did not have the four other glycosylation sites, namely, 44 (NHT), 70 (NNT), 434 (NTT), and 455 (NWS); this finding indicates that the overall spiral structure of A/Beijing/3/2009 (H1N1) did not change, unlike that of the A/Taiwan/5072/1999 (H1N1) strain (Figure 2).
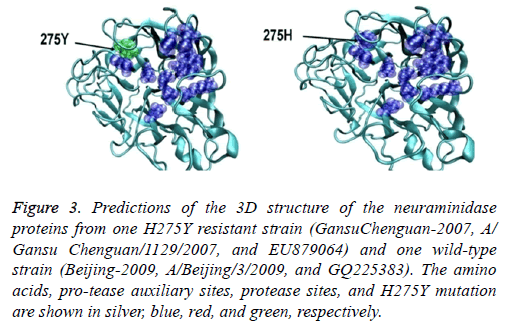
The eight sialidase activity sites and 11 auxiliary centers were consistent and had a pocket-like structure. This structure was located on the surface of the internal H275Y resistance site locus. No obvious difference in the structure of the sialidase auxiliary and sialidase centers was observed between the H275Y resistant (A/Gansu/Chenguan/1129/2007 (H1N1)) and wild-type strains (A/Nanchang/4/1998 (H1N1)) or between the novel 2009 resistant (A/Hong Kong/17566/2009 (H1N1)) and wild-type strains (A/Beijing/3/2009 (H1N1)). However, the 2009 novel and resistant strains had clear structural differences (Figure 3).
Figure 3: Predictions of the 3D structure of the neuraminidase proteins from one H275Y resistant strain (GansuChenguan-2007, A/ Gansu Chenguan/1129/2007, and EU879064) and one wild-type strain (Beijing-2009, A/Beijing/3/2009, and GQ225383). The amino acids, pro-tease auxiliary sites, protease sites, and H275Y mutation are shown in silver, blue, red, and green, respectively.
Discussion
This study describes the evolution of NA genes over several years in different countries. The results show that the new 2009 H1N1 influenza virus is 99.9% homologous across different areas but only 75% homologous to the 1995-2008 H1N1 influenza virus. Therefore, the 2009 pandemic H1N1 influenza virus was caused by a rearrangement of the virus that evolved from a completely different ancestor rather than from the previous species circulating worldwide. This finding corresponds to the lack of immunity to the 2009 H1N1 influenza A virus observed among young people who were infected [4,15].
In this study, many amino acid substitutions existed in the AD sites. Approximately 20% of the NA substitutions were distributed in the following sites: aa23, aa45, aa78, aa79, aa80, aa81, aa84, aa149, aa222, aa234, aa241, aa248, aa274, aa287, aa321, aa344, aa427, aa430, aa432, aa436, aa454, and aa455. The most common NA AD substitutions in China during 1995-2012 were M23I (38%, 76/200), R222N (37%, 74/200), N248D (38.5%, 77/200), V321I (41.5%, 83/200), D344N (40.5%, 81/200), L430R (39.5%, 79/200), and R432K (39.5%, 79/200). Sites aa23, aa45, aa78, and aa79 are located in the stalk region of NA, and their length affects the host range and replication of influenza A viruses [4,16,17]. All of the novel 2009 strains had H45Q substitution. Luo [17] reported that the stalk region of NA is essential for the infectious virus, particularly site 76 because of its proximity to 78 and 79. Changes in I117 and E119 alter the susceptibility to NA inhibitors [18]. Additionally, an analysis of the isolates between 2006 and early 2008 from Australasia and Southeast Asia has revealed that aa136 substitution can reduce the susceptibility of the virus to zanamivir.
N248D substitution can change the central region of the antibody recognition site [14], and whether changes in these three sites (222, 249, and 344), which are in the vicinity of the catalytic site, affect the catalytic site and efficiency of the enzyme remains unknown. V321I dis-torts the hydrophobic pockets and affects residues in the NA active site [19].
Moreover, the NA amino acid region between 426 and 457 is essential for stability maintenance at a low pH and that alterations at positions 430, 432, 434, and 455 (N1 numbering) or deletion at position 435 in the NA can affect stability at a low pH. Increased stability at a low pH allows the influenza virus to replicate rapidly [20]. Overall, some of the mutation sites in NA clearly influence its structure and function. The virus undergoes frequent point mutations in response to drugs and immune system pressure. Thus, further studies on the effect of the virulence of these sites and whether these sites are related to the pandemic influenza H1N1 in different years may help guide the monitoring of new species and changes in virulence and transmissibility.
Nineteen NA sialidase active and auxiliary center sites were conserved in different years at different countries. However, we observed several strains that contained the H275Y substation. These sites are highly conserved because they are directly or indirectly involved in the activity of the enzyme. H275Y is the most widely acknowledged substitution that is responsible for resistance to oseltamivir; the mutant reduces the affinity for the substrates compared with species lacking the substitution, although it has no effect on susceptibility to zanamivir or adamantine [21,22]. The mutation rate of H275Y in our research was 5.5% (11/200). In China, this substitution was observed in only one strain, that is, A/Gansu/Chenguan/ 1129/2007, which was isolated before 2008. However, five novel H1N1 strains isolated from Hong Kong and Taiwan during 2009-2012 were found to carry this mutation. Amino acid 275, which is located on the surface of NA, is important for interaction with antiviral drugs. The H275Y mutation did not change the 3D structure of the NA protein. The NA amino acid substitutions may have affected the drug resistance to NA inhibitors. Studies have shown that the H275Y mutant causes 1,466-fold reduced sensitivity to Tamiflu [21,23]. The other site mutations, such as E119K and D151N, do not have a clear relationship to drug sensitivity because they are observed at a low frequency. Although the influenza virus resource reported that Tamiflu-resistant strains have been found in Denmark, Japan, Hong Kong, and China [24,25], their proportion is approximately 11.55% (1226/10612), which would not significantly affect public health and safety. However, the seasonal influenza and the new influenza (H1N1) co-circulate in the population and can significantly increase the risk of a recombinant strain emerging. Therefore, changes in the virus should be closely monitored, and the occurrence and spread of resistant strains should be pre-vented.
The distribution of the glycosylation sites is consistent with that in the new 2009 influenza A H1N1. The sites are located at 88 NSS, 146 NGT, and 235 NGS. Compared with the seasonal H1N1 prior to 2009, four sites, namely, 44 (NHT), 70 (NNT), 434 (NTT), and 455 (NWS), and three additional sites, namely, 50 (NQS), 58 (NNT), and 63 (NQT), were observed. Glycosylation sites 88, 146, 235, and 455 are located on the surface of the protein, and they are related to host adaptation, transmissibility, and pathogenicity. These highly conserved glyco-sylation sites are important for the NA protein to maintain its structure and physiological function [26]. Many studies both in silico or through actual experiments have indicated that an increase or decrease in glycosylation sites or glycosylation migration influences receptor binding, antigenic site blocking, transfaunation, pathogenicity and so on [13,27-32]. However, further studies on the possible role of glycosyl modification of NA are required.
Conclusion
In conclusion, the evolutionary features of the Chinese H1N1 NA gene are as follows.
(1) The 2009 pandemic influenza A/H1N1 NA genes were similar in viruses in different countries worldwide and varied in the seasonal virus prior to 2009.
(2) The strains isolated during 1995-2008 had seven highly conserved glycosylation sites, with the exception of 44 (NHT), 50 (NQS), and 70 (NNT). The strains from 2009 had four sites only, of which three sites (88 NSS, 146 NGT, and 235 NGS) were identical to the seasonal H1N1 gene. This similarity indicates that these three stable sites are important for the virus’s structure. Additionally, three sites (50 NQS, 58 NST, and 63 NHT) were different. Four sites (44 NHT, 434 NTT, 455 NWS, and 70 NNT) were unimportant for adaptation to the host, transmissibility, or pathogenicity. Overall, 19 NA sialidase activity and auxiliary center sites were highly conserved over the past 18 y. H275Y is the most widely acknowledged substitution that is responsible for resistance to oseltamivir, but its prevalence rate is very low, and it does not pose a threat to public health.
(3) With a mutation frequency of more than 20% focused on the antigenic sites in the stem and catalytic site of the virus (23, 45, 78, 79, 80, 81, 84, 149, 222, 234, 248, 274, 321, 344, 427, 430, 432, 436, 454, and 455), whether the antigenic sites will enhance virulence, epidemic range, or other features remains unclear.
(4) No differences were observed in potential glycosylation sites and NA sialidase activities (except for H275Y) among the NA-resistant strains from China and other countries and the wild-type strains prior to 2009. The NA resistant strains from 1995-2008 had only 80%-81% similarity to the 2009 wild-type and resistant viruses.
This study described the genetic evolution of NA from human influenza A/H1N1 viruses during 1995-2012 in China. The results are expected to be beneficial for early warning and laboratory diagnosis of influenza H1N1 pandemics. Although H1N1 influenza appeared in the low epidemic period, it still underwent “antigenic shift” and “antigenic drift” for seasonal and novel co-circulation in the population. Therefore, virological surveillance, particularly the monitoring of Tamiflu-resistant strains and molecular evolution of key sites, should be strengthened.
References
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev 1992; 56: 152-179.
- Liu SL, Zhang ZR, Wang C, Dong Y, Cui LB, Yang XH. 2009 pandemic characteristics and controlling experiences of influenza H1N1 virus 1 year after the inception in Hangzhou, China. J Med Virol 2010; 82: 1985-1995.
- Liu SL, Zhang Z, Wang C, Dong Y, Pan H. Evolutionary characterization of human H1N1 influenza virus hemagglutinin genes isolated from 1947 to 2009 in China. Intervirology 2011; 54: 233-245.
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A. Antigenic and genetic characteristics of swine-origin 2009 A (H1N1) influenza viruses circulating in humans. Science 2009; 325: 197-201.
- Grienke U, Schmidtke M, von Grafenstein S, Kirchmair J, Liedl KR. Influenza neuraminidase: a druggable target for natural products. Nat Prod Rep 2012; 29: 11-36.
- Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 2006; 443: 45-49.
- Hurt AC, Ernest J, Deng Y-M, Iannello P, Besselaar TG, Birch C. Emergence and spread of oseltamivir-resistant A (H1N1) influenza viruses in Oceania, South East Asia and South Africa. Antiviral Res 2009; 83: 90-93.
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-Model: an automated protein homology modeling server. Nucleic Acids Res 2003; 31: 3381-3385.
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph 1996; 14: 33-38, 27-28.
- Shtyrya YA, Mochalova LV, Bovin NV. Influenza virus neuraminidase: structure and function. Acta Naturae 2009; 1: 26-32.
- Sheu TG, Deyde VM, Okomo-Adhiambo M, Garten RJ, Xu X, Bright RA. Surveil-lance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob Agents Chemother 2008; 52: 3284-3292.
- Yen HL, Ilyushina NA, Salomon R, Hoffmann E, Webster RG, Govorkova EA. Neu-raminidase inhibitor-resistant recombinant A/Vietnam/1203/04 (H5N1) influenza viruses retain their replication efficiency and pathogenicity in vitro and in vivo. J Virol 2007; 81: 12418-12426.
- Chen W, Zhong Y, Qin Y, Sun S, Li Z. The evolutionary pattern of glycosylation sites in influenza virus (H5N1) hemagglutinin and neuraminidase. PloS one 2012; 7: 49224.
- Maurer-Stroh S, Ma J, Lee RT, Sirota FL, Eisenhaber F. Mapping the sequence mutations of the 2009 H1N1 influenza A virus neuraminidase relative to drug and antibody binding sites. Biol Direct 2009; 4: 18.
- Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE Jr. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 2010; 328: 357-360.
- Guangxiang L, Jeffrey C, Palese P. Alterations of the stalk of the influenza virus neuram-inidase: deletions and insertions. Virus Res 1993; 29: 141-153.
- Castrucci MR, Kawaoka Y. Biologic importance of neuraminidase stalk length in influenza A virus. J Virol 1993; 67: 759-764.
- Deyde VM, Sheu TG, Trujillo AA, Okomo-Adhiambo M, Garten R, Klimov AI. Detection of molecular markers of drug resistance in 2009 pandemic influenza A (H1N1) vi-ruses by pyrosequencing. Antimicrob Agents Chemother 2010; 54: 1102-1110.
- Stoner TD, Krauss S, Dubois RM, Negovetich NJ, Stallknecht DE. Antiviral susceptibility of avian and swine influenza virus of the N1 neuraminidase subtype. J Virol 2010; 84: 9800-9809.
- Takahashi T, Kurebayashi Y, Ikeya K, Mizuno T, Fukushima K, Kawamoto H. The low-pH stability discovered in neuraminidase of 1918 pandemic influenza A virus enhances virus replication. PLoS One 2010; 5: 15556.
- Li Q, Qi J, Zhang W, Vavricka CJ, Shi Y. The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site. Nat Struct Mol Biol 2010; 17: 1266-1268.
- Hurt AC, Holien JK, Parker M, Kelso A, Barr IG. Zanamivir resistant influenza viruses with a novel neuraminidase mutation. J virol 2009; 83: 10366-10373.
- Ferraris O, Lina B. Mutations of neuraminidase implicated in neuraminidase inhibitors resistance. J Clin Virol 2008; 41: 13-19.
- Gooskens J, Jonges M, Claas EC, Meijer A, van den Broek PJ, Kroes AC. Morbidity and mortality associated with nosocomial transmission of oseltamivir-resistant influenza A (H1N1) virus. JAMA 2009; 301: 1042-1046.
- Dharan NJ, Gubareva LV, Meyer JJ, Okomo-Adhiambo M, McClinton RC, Marshall SA. Infections with oseltamivir-resistant influenza A (H1N1) virus in the United States. JAMA 2009; 301: 1034-1041.
- Sun S, Wang Q, Zhao F, Chen W, Li Z. Glycosylation site alteration in the evolution of influenza A (H1N1) viruses. PLoS One 2011; 6: 22844.
- Wu ZL, Ethen C, Hickey GE, Jiang W. Active 1918 pandemic flu viral neuraminidase has distinct N-glycan profile and is resistant to trypsin digestion. Biochem Biophys Res Commun 2009; 379: 749-753.
- Sun S, Wang Q, Zhao F, Chen W, Li Z. Prediction of biological functions on glycosylation site migrations in human influenza H1N1 viruses. PLoS One 2012; 7: 32119.
- Li S, Schulman J, Itamura S, Palese P. Glycosylation of neuraminidase determines the neurovirulence of influenza A/WSN/33 virus. J Virol 1993; 67: 6667-6673.
- Tisoncik JR, Guo Y, Cordero KS, Yu J, Wang J, Cao Y. Identification of critical residues of influenza neuraminidase in viral particle release. Virol J 2011; 8: 1.