ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Review Article - Biomedical Research (2021) Biomedical and cellular systems for diagnostic and therapeutic purposes
Effect of UGT2B7 Gene Polymorphism with Clinical Pediatric Epileptic patients on Sodium Valproate Monotherapy
1K S Hegde Medical Academy, Nitte (Deemed to be University), Mangalore, Karnataka, India
2Departmentt of Pharmacology, Biochemistry, Pediatrics and Genetics, K S Hegde Medical Academy, Nitte (Deemed to be University), Mangalore, Karnataka, India
- Corresponding Author:
- Sachidananda Adiga
Department of Pharmacology
K S Hegde Medical Academy
Mangalore
Karnataka
India
Accepted January 12, 2021
Background: Pediatric epilepsy comprises of a chronic neurological disorders characterized by recurrent seizure attack. Sodium valproate is one of the common anti-epileptic drugs used in the treatment. Glucuronide conjugation is the major metabolic pathway of sodium valproate, carried out by the enzyme Uridine 5’-diphospho (UDP) glucuronosyl transferase (UGT) whose genetic polymorphisms may alter clinical outcome. Aim: To find the association between UGT2B7 genetic polymorphism and clinical outcome in terms of efficacy and tolerability in pediatric epileptic patients on sodium valproate monotherapy. Methods and Materials: In this cohort study, 75 pediatric epileptic patients aged 2-18 years receiving sodium valproate monotherapy for past one month were included from Justice K S Hegde Charitable Hospital, Mangalore, India after obtaining informed consent. Genetic polymorphism patterns of UGT2B7 (C161T, A268G, G211T) was evaluated by PCR-RFLP. Clinical outcome was measured in terms of responders and non-responders based on seizure control during the 6 month observation period. Tolerability was measured by estimating the hepatic, renal and other lab parameters. Clinical outcome in different UGT genotypes was compared by Chi square test. P value <0.05 was considered as significant. Results: Out of 75 patients, CC (41.3%), CT (38.7%), TT (20%) pattern was observed in UGT2B7 (C161T) gene, AA(14.7%), AG(42.7%), GG(42.7%) in (A268G) gene and GG(80%), GT(18.7%), TT(1.3%) in (G211T) gene. It was found that there was no statistical difference in clinical outcome with different UGT2B7 genetic polymorphism patterns. Conclusion: We conclude from our study that genetic polymorphism of UGT2B7 doesn’t have any role on the clinical outcome of epilepsy.
Keywords
UGT2B7, Genetic polymorphism, Sodium valproate, Clinical Outcome.
Introduction
Epilepsy is a neurological disorder comprising of heterogeneous and complex brain disorder of many seizure types and epilepsy syndromes [1,2]. The overall prevalence of epilepsy in India is estimated to be 3-11.9 per 1000 population [3]. It is common practice for pediatricians and neurologists to begin long-term, daily Anti-Epileptic Drugs (AED) after a patient has experienced unprovoked seizures [4]. The AEDs lead to satisfactory seizures control for about 60–70% of newly treated patients [5] while remaining 30% of patients will have uncontrolled epilepsy with AED treatment for prolonged period with recurrent seizures, adverse effects and significantly increased risk of mortality and morbidity [1,5]. The goal of treatment of epilepsy in pediatric patient is to prevent the seizure attack for two years with an appropriate Anti-Epileptic Drug (AED) with minimal adverse effects so that the drug can be slowly tapered off later.
Pharmacogenetics refers to the science about the genetic variations affecting drug metabolism, drug targets or disease pathways leading to varying response to the drug with regard to its efficacy or adverse effect. Hence it is important to investigate the role of genetic polymorphism that can alter drug metabolism and effect of antiepileptic drugs. Sodium valproate is a widely used broad spectrum antiepileptic drug due to its effect on different types of epilepsy. Glucuronidation and β-oxidation are the prominent metabolic pathways, while CYP-mediated oxidation is a minor pathway in metabolizing of Sodium Valproate drug. Glucuronidation is catalyzed by Uridine- 5´-diphosphate Glucuronosyl Transferases (UGTs), Polymorphisms affecting genes encoding for UGT enzymes may result in alterations of sodium valproate metabolism. Changes in the glucuronidation rate of sodium valproate may alter the blood level of drug resulting in either insufficient plasma concentration that leads to reduced effectiveness of therapy or elevated plasma concentration of drug that manifests as toxicity [6]. Previous researchers have focused on UGT2B7 polymorphisms in Chinese and Japanese population. Most of the studies have aimed at finding the pattern of genetic polymorphism and its effect on serum concentration of patient. However, currently available information of sodium valproate on UGT2B7 polymorphism and its association with clinical outcome in Indian population is very limited.
Objective
To find association of UGT2B7 gene polymorphism with clinical outcome in terms of efficacy and tolerability in pediatric epileptic patients on sodium valproate monotherapy.
Methodology
In this cohort study, 75 pediatric epileptic patients of South Indian origin were enrolled, which was carried out at pediatric department of Justice K S Hegde Charitable Hospital, Mangalore, India and Central Research Laboratory of K S Hegde Medical Academy, Mangalore from February 2018 to December 2019. Children aged 2-18 years of either sex with diagnosis of epilepsy treated with sodium valproate monotherapy were enrolled. Ethics Committee approval was obtained prior to the study. Written informed consent and ascent were obtained prior to enrollment. Patients diagnosed clinically, by EEG and who are on steady stable dose of 20mg/kg/day sodium valproate twice/thrice a day for past one month was included. Patients having history or evidence of hepatitis or impaired renal functions or who are already on treatment with any other antiepileptic drugs or drugs which induce or inhibit sodium valproate metabolism were excluded.
Four ml of whole blood sample was collected with aseptic precaution once patient fulfilled criteria and during 6month follow-up visit. The sample was separated into 2 parts; 2 ml of EDTA whole blood stored at -80ºC for genetic polymorphism analysis, 2 ml of whole blood in plain vial centrifuged to obtain serum and then stored at -80ºC to analyze biochemical parameters. Liver function test (Albumin, total protein, total, direct bilirubin, SGOT, SGPT, ALP), renal function test (Blood urea, serum creatinine), platelet count and serum amylase were estimated at the time of enrollment and at 6 month.
Genotyping
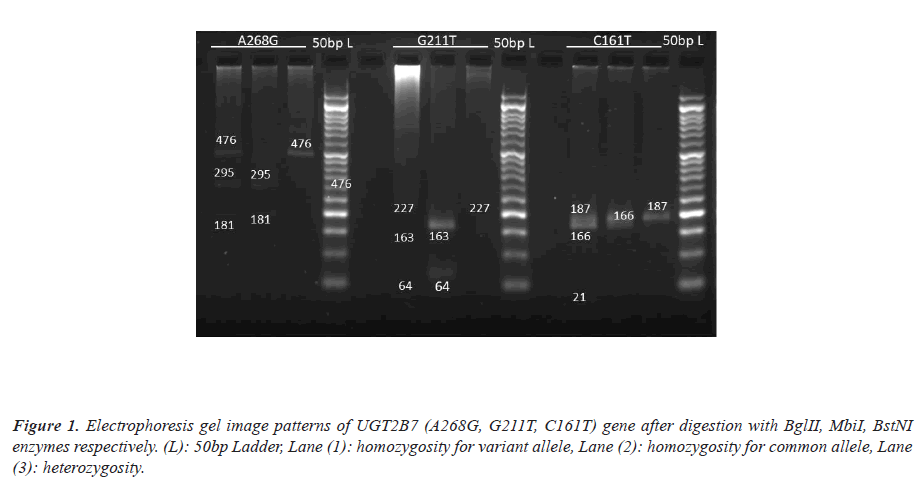
Genomic DNA from whole blood was isolated by Phenolchloroform extraction and ethanol precipitation method. The UGT2B7 (C161T, A268G, G211T) polymorphisms was analyzed by the Polymerase Chain Reaction–Restriction Fragment Length Polymorphism (PCR–RFLP) method [7,8]. Amplification was performed in MJ-Mini Thermal cycler (Bio-Rad, Tokyo, Japan). PCR was conducted with an initial denaturation enzyme activation step at 95°C for 5 minutes, amplification step for 35 cycles at 95°C for 30 seconds, an annealing temperature for 30 seconds, treatment at 72°C for 30 seconds and a final extension step at 72°C for 5 minutes. PCR product was digested with each restriction enzyme (BstNI, BglII, MbiI) respectively for UGT2B7 (C161T, A268G, G211T). Amplified product of DNA samples and restriction fragments was separated on a 2% agarose gel with ethidium bromide.
The genotype was assigned based on the results of the analysis of the digestion patterns.
Parents were educated to maintain a diary in which they were instructed to record seizure attack if any, duration of attack, change in the pattern from previous episode. They were also instructed to record the adherence to medication by recording the days the child had skipped the medication. They were also instructed to bring the child for clinical visit in the event of nausea, vomiting, loss of appetite, jaundice, pain abdomen anytime or otherwise at the end of 6 month.
Clinical outcome was assessed in terms of efficacy and safety to sodium valproate. Efficacy of the drug was measured in terms of treatment responders or nonresponders. Responder to sodium valproate was defined as patients with no relapse or <2 episodes of seizure attack in 6 consecutive months of follow up and is well tolerated with sodium valproate monotherapy without any adverse effect seen. Non-responders were those with two or more than two episode of seizure attack in six months of follow up or with any adverse drug effect seen after therapy initiation [9]. Tolerability was assessment by comparing the biochemical parameters between basal and at six month interval. Tolerability was also assessed whether any patients experience jaundice, pancreatitis or signs of thrombocytopenia.
Sample size calculation
Based on the genotype frequencies reported previously and taking drug concentration SD as 23μg/ml [10] and fixing the margin of error as 5, the calculated sample size was 87. However due to the change in treatment guidelines of pediatric epilepsy given by Indian Pediatric Association (preference of levetiracetam over sodium valproate), we could enroll only 65 patients.
Statistical analysis
The collected data was expressed as frequency, percentage for qualitative data, and mean ± SD for quantitative data using SPSS software (version 16.0). Comparison between responders and non-responders in different gene patterns of UGT 2B7 studied was analyzed by using Chi square test. Biochemical parameters were compared between basal and 6 months follows up by paired‘t’ test between different age groups. The P value <0.05 was considered as statistically significant.
Results
The mean age of patient enrolled in the study was 7.38± 3.67 yrs. There were 47 male and 28 female patients in our study and the mean body mass index was 16.26± 5.32. Out of 75 patients, CC (41.3%), CT (38.7%), TT (20%) pattern was observed in UGT2B7 (C161T) gene, AA(14.7%), AG(42.7%), GG(42.7%) in (A268G) gene and GG(80%), GT(18.7%), TT(1.3%) in (G211T) gene (Figure 1).
The frequencies were consistent with the Hardy-Weinberg equilibrium (Table 1).It was observed that there was no statistical difference in clinical outcome with UGT2B7 (C161T, A268G, G211T) genetic polymorphism (Table 2). It was also observed that there was higher pattern of mutant carrier alleles for the genes UGT2B7 (C161T and A268G). All the children had normal liver function parameters, renal function parameters during basal and 6 months follow up (Table 3). One patient developed acute pancreatitis, later drug was discontinued and patient was put on levetiracetam. There was no statistical difference of drug concentration observed in different UGT2B7 gene polymorphism (Table 4).
| Genotype | Actual Values (%) | Expected Values (%) | χ2 |
|---|---|---|---|
| C161T | |||
| CC | 38.4 | 35.52 | 1.429 |
| CT | 42.4 | 48.15 | |
| TT | 19.2 | 16.32 | |
| A268G | |||
| AA | 15.2 | 12.18 | 1.766 |
| AG | 39.4 | 45.44 | |
| GG | 45.4 | 42.38 | |
| G211T | |||
| GG | 80.8 | 79.92 | 0.855 |
| GT | 17.2 | 18.95 | |
| TT | 02 | 1.12 | |
Table 1. Hardy Weinberg Equilibrium test for UGT2B7 gene.
| Genotype UGT2B7 |
Polymorphism pattern | Polymorphism in percentage | Clinical Outcome | P Value | |
|---|---|---|---|---|---|
| Responders (N=57) | Non- responders (N=18) |
||||
| C161T | CC | 41.3% | 24 22 11 |
07 07 04 |
0.965 |
| CT | 38.7% | ||||
| TT | 20% | ||||
| A268G | AA | 14.7% | 09 24 24 |
02 08 08 |
0.887 |
| AG | 42.7% | ||||
| GG | 42.7% | ||||
| G211T | GG | 80% | 48 09 00 |
12 05 01 |
0.093 |
| GT | 18.7% | ||||
| TT | 1.3% | ||||
Statistical test used is Chi Square test
Table 2. Association of UGT2B7 gene polymorphisms with Clinical Outcome (N=75).
| UGT2B7 (C161T) | UGT2B7 (A268G) | UGT2B7 (G211T) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | AA | AG | GG | GG | GT | TT | ||
| Alb (g/dl) |
Basal 6 months P value |
4.4 ± 0.4 4.5 ± 0.3 0.369 |
4.5 ± 0.3 4.5 ± 0.3 0.938 |
4.5 ± 0.3 4.5 ± 0.5 0.884 |
4.6 ± 0.2 4.5 ± 0.3 0.127 |
4.5 ± 0.3 4.4 ± 0.4 0.269 |
4.4 ± 0.4 4.6 ± 0.3 0.052 |
4.4 ± 0.3 4.5 ± 0.3 0.359 |
4.5 ± 0.3 4.4 ± 0.4 0.647 |
---- |
| Tot Prot (g/dl) | Basal 6 months P value |
7.4 ± 0.2 7.6 ± 0.1 0.232 |
7.4 ± 0.1 7.4 ± 0.2 0.594 |
7.4 ± 0.1 7.5 ± 0.2 0.790 |
7.3 ± 0.6 7.7 ± 0.7 0.142 |
7.2 ± 0.4 7.3 ± 0.6 0.177 |
7.5 ± 0.9 7.7 ± 0.6 0.481 |
7.3 ± 0.6 7.6 ± 0.6 0.038 |
7.5 ± 0.8 7.4 ± 0.7 0.736 |
---- |
| SGOT (IU/L) | Basal 6 months P value |
28.1 ± 9.5 27.5 ± 9.2 0.818 |
35.7 ± 29 27.5 ± 9 0.232 |
27.4 ± 10 26.9 ± 9 0.766 |
29.5 ± 9.4 26.5 ± 3.8 0.334 |
27 ± 7.1 27.1 ± 7.6 0.873 |
347 ± 29.1 27.3 ± 11.3 0.258 |
30.4 ± 20.4 26.8 ± 8.9 0.229 |
30 ± 8.4 29 ± 7.4 0.527 |
---- |
| SGPT (IU/L) | Basal 6 months P value |
11.4 ± 3.7 13.6 ± 6.7 0.120 |
13.8 ± 5.2 12.5 ± 5.0 0.420 |
14.4 ± 6.1 12.6 ± 4.8 0.205 |
13.8 ± 3.6 12 ± 4.7 0.434 |
11.2 ± 4.6 12.2 ± 3.9 0.256 |
13.6 ± 5.4 13.7 ± 7.5 0.963 |
12.5 ± 4.6 12.4 ± 5.6 0.890 |
12.8 ± 6.2 14.6 ± 5.2 0.345 |
---- |
| ALP (IU/L) |
Basal 6 months P value |
188 ± 64 209 ± 89 0.289 |
216 ± 66 202 ± 70 0.329 |
209 ± 100 217 ± 122 0.860 |
237 ± 56 226 ± 76 0.640 |
189 ± 61 199 ± 86 0.512 |
192 ± 85 201 ± 90 0.706 |
197 ± 73 206 ± 89 0.513 |
205 ± 68 197 ± 67 0.640 |
---- |
| BID (mg/dl) |
Basal 6 months P value |
0.12 ± 0.19 0.09 ± 0.04 0.466 |
0.13 ± 0.11 0.12 ± 0.04 0.556 |
0.09 ± 0.04 0.12 ± 0.07 0.250 |
0.09 ± 0.04 0.08 ± 0.03 0.209 |
0.09 ± 0.04 0.11 ± 0.05 0.061 |
0.14 ± 0.21 0.11 ± 0.05 0.393 |
0.09 ± 0.08 0.10 ± 0.05 0.394 |
0.2 ± 0.28 0.11 ± 0.05 0.282 |
---- |
| BIT (mg/dl) |
Basal 6 months P value |
0.33 ± 0.27 0.26 ± 0.14 0.125 |
0.35 ± 0.2 0.32 ± 0.2 0.288 |
0.27 ± 0.11 0.34 ± 0.21 0.305 |
0.27 ± 0.1 0.22 ± 0.1 0.061 |
0.27 ± 0.1 0.31 ± 0.2 0.178 |
0.38 ± 0.3 0.30 ± 0.2 0.173 |
0.29 ± 0.17 0.29 ± 0.16 0.899 |
0.44 ± 0.34 0.31 ± 0.16 0.102 |
---- |
| Urea (mg/dl) |
Basal 6 months P value |
20.8 ± 6.9 20.4 ± 8.5 0.775 |
22.1 ± 6.9 19.5 ± 7.1 0.201 |
24.1 ± 9.5 17.5 ± 6.7 0.098 |
20.8 ± 5.9 20.5 ± 8.8 0.919 |
21.9 ± 8.5 19.3 ± 8.0 0.177 |
20.9 ± 7.1 19.5 ± 7.4 0.355 |
21.4 ± 7.3 19.3 ± 8.2 0.093 |
21.1 ± 9.2 20.9 ± 5.1 0.959 |
---- |
| Creat (mg/dl) |
Basal 6 months P value |
0.43 ± 0.23 0.38 ± 0.15 0.272 |
0.55 ± 0.2 0.41 ± 0.1 0.002* |
0.4 ± 0.2 0.31 ± 0.13 0.062 |
0.53 ± 0.21 0.42 ± 0.1 0.116 |
0.49 ± 0.2 0.37 ± 0.12 0.00* |
0.46 ± 0.3 0.39 ± 0.2 0.111 |
0.49 ± 0.23 0.38 ± 0.13 0.000* |
0.43 ± 0.15 0.42 ± 0.11 0.636 |
---- |
| Amylase (IU/L) |
Basal 6 months P value |
79.4 ± 29.5 81.6 ± 30.7 0.755 |
85.4 ± 34 84.1 ± 35 0.837 |
88.9 ± 44.4 86.4 ± 38.8 0.812 |
93.5 ± 49 101 ± 48 0.478 |
94.9 ± 53 83.1 ± 28 0.176 |
75.7 ± 27.3 77.5 ± 32.2 0.797 |
85.7 ± 45 83.7 ± 32.8 0.717 |
96.1 ± 42.5 84.2 ± 38.5 0.241 |
---- |
| Platelets (lakh cells/cu mm) |
Basal 6 months P value |
2.69 ± 0.74 2.68 ± 0.43 0.990 |
2.61 ± 0.51 2.77 ± 0.43 0.196 |
3.03 ± 0.60 2.73 ± 0.43 0.183 |
2.92 ± 0.63 2.67 ± 0.50 0.247 |
2.62 ± 0.57 2.74 ± 0.42 0.341 |
2.68 ± 0.68 2.73 ± 0.42 0.687 |
2.72 ± 0.63 2.77 ± 0.40 0.558 |
2.54 ± 0.60 2.49 ± 0.50 0.839 |
---- |
Statistical test used is paired ‘t’test
Table 3. Association of biochemical parameters between basal and 6 months with different gene pattern. (N=75).
| Genotype UGT2B7 |
SV Concentration (µg/ml) | SV dose (mg/kg) | Adjusted SV Concentration [(µg/ml)/(mg/kg)] |
|---|---|---|---|
| C161T CC (38.4%) CT (42.4%) TT (19.2%) (P value) |
106.0 ± 37.8 95.9 ± 37.7 105.8 ± 17.2 (0.939) |
20.6 ±6.6 21.5 ±6.7 24.3 ±13.6 (0.091) |
5.51 ± 2.6 4.90 ± 2.7 5.71 ± 3.3 (0.380) |
| A268G AA (15.2%) AG (39.4%) GG (45.4%) (P value) |
117.5 ± 35.7 97.4 ± 32.8 101.4 ± 35.6 (0.831) |
22.6 ± 8.1 22.0 ± 7.9 21.1 ±9.2 (0.825) |
5.78 ± 3.3 5.16 ± 2.9 5.36 ± 2.6 (0.737) |
| G211T GG (80.8%) GT (17.2%) TT (02%) (P value) |
99.7 ± 36.5 110.8 ± 24.9 123 (0.503) |
21.6 ± 7.2 19.6 ± 8.8 25.5 (0.668) |
5.03 ± 2.5 6.79 ± 3.5 4.82 (0.178) |
One-way ANOVA followed by Bonferroni’s post hoc test. *P value <0.05 was statistically significant
Table 4. Comparison of UGT2B7gene polymorphisms with Sodium valproate (SV) concentration and Adjusted SV Concentration (N=75).
Discussion
In the present study we have tested the associations between gene pattern of UGT2B7 gene with clinical outcome. Gene polymorphism might play an important role in the interindividual variability in pharmacokinetics and pharmacodynamics in pediatric epileptic patients on sodium valproate monotherapy. Many researchers have investigated role of gene polymorphisms in pharmacokinetics and pharmacodynamics aspects of sodium valproate. The pattern of UGT2B7 gene polymorphism was similar when compared to the Chinese and Japanese population with respect to UGT2B7 (C161T, A268G, G211T type) (Table 5) [7,8,10-12 ]. In Chinese pediatric epileptic population [10], the predominant genetic polymorphism of UGT2B7 (C161T) gene pattern observed was wild type while in ours and Japanese study population the predominant pattern of UGT2B7 (C161T) observed was mutant carrier allele type. However there was no difference in genetic pattern observed between our patients and Japanese pediatric epileptic patients [7].
| Genotype UGT2B7 | Chinese patients with epilepsy | Japanese | Current Study | |||
|---|---|---|---|---|---|---|
| Hongying et al. (n=162) | Zhongliang et al. (n=102) | Hung et al. (n=162) | Sun et al. (n=102) | Inoue et al. (n=78) | (n=99) | |
| C161T CC CT TT | NS NS NS | NS NS NS | 55.56% 37.04% 07.41% | NS NS NS | 37.2% 53.8% 09.0% | 38.4% 42.4% 19.2% |
| A268G AA AG GG | 05.65% 48.79% 45.56% | 05.88% 49.01% 45.11% | NS NS NS | NS NS NS | NS NS NS | 15.2% 39.4% 45.4% |
| G211T GG GT TT | 59.27% 36.29% 04.44% | 72.55% 25.50% 01.95% | NS NS NS | 76.47% 22.55% 0.98% | NS NS NS | 80.8% 17.2% 02.0% |
NS: Not Studied
Table 5. Details of prevalence of UGT2B7 gene polymorphisms in various populations.
Different studies conducted in Chinese pediatric epileptic were similar to our study population i.e UGT2B7 (A268G) had higher population of mutant carrier allele type when compared to wild type and UGT2B7 (G211T) had lower mutant carrier allele type carrier when compared to wild type.
To date, reports on the influence of UGT2B7 polymorphism on substrate drug metabolism still remain inconsistent, Guo et al. [13] reported that there was no significant effect of polymorphisms on serum valproate concentrations. However Hongying et al and Zhongliang et al. [8,11] reported that UGT2B7 (A268G) plays an important role in sodium valproate metabolism i.e Wild type had higher drug concentration when compared to mutant carrier allele type which was statistically significant. Inoue et al. [7] reported that patients with UGT2B7 (C161T) genotype had lower adjusted plasma valproic acid concentration when compared to mutant carrier allele type. None of the available literatures till date have explained the influence of UGT genetic variants on clinical outcome of sodium valproate monotherapy. It was tentatively approached by Hung et al. who explained that patients showing good drug response (seizure free/good seizure control) to evaluate multiple genetic influence including UGT1A6 gene on VPA pharmacokinetics [10]. However in our study we could not find any association between different pattern of polymorphism with clinical efficacy and tolerability.
As per literature survey, there is no published data on comparison of UGT2B7 gene polymorphism with clinical outcome and safety parameters in Indian pediatric epileptic population. Hence we made an attempt to find the association of UGT2B7 gene polymorphism among pediatric epileptic patients with clinical efficacy and tolerability of sodium valproate.
The limitation of our study was smaller sample size. In the present context of changed protocol in the treatment of pediatric epilepsy (preference of levetiracetam over sodium valproate) only metacentric study involving different region may answer our limitation and add quality to the data in terms genetic pattern.
Conclusion
We conclude from our study that different patterns of UGT2B7 genetic polymorphism don’t have any association on the clinical outcome of Sodium valproate in terms of efficacy and tolerability of epilepsy. We also conclude that sodium valproate was well tolerated among the pediatric patients with epilepsy and can be used an effective anti-epileptic drug.
References
- Tejada J, Costa KM, Bertti P, Garcia-Cairasco N. The epilepsies: complex challenges needing complex solutions. Epilepsy Behav 2013; 26: 212-228.
- Berg AT, Berkovic SF, Brodie MJ, Buchhalter J, Cross JH, van Emde Boas W. Revised terminology and concepts for organization of seizures and epilepsies: report of the ILAE Commission on Classification and Terminology, 2005-2009. Epilepsia 2010; 51: 676-685.
- Amudhan S, Gururaj G, Satishchandra P. Epilepsy in India I: epidemiology and public health. Ann Indian Acad Neurol 2015; 18: 263-277.
- Cockerell OC, Johnson AL, Sander JW, Shorvon SD. Prognosis of epilepsy: a review and further analysis of the first nine years of the British National General Practice Study of Epilepsy, a prospective population-based study. Epilepsia 1997; 38: 31-46.
- Sillanpaa M, Schmidt D. Natural history of treated childhood-onset epilepsy: prospective, long-term population-based study. Brain 2006; 129: 617-624.
- Chatzistefanidis D, Georgiou I, Kyritsis AP, Markoula S. Functional impact and prevalence of polymorphisms involved in the hepatic glucuronidation of valproic acid. Pharmacogenomics 2012; 13: 1055-1071.
- Inoue K, Suzuki BS, Yazawa R, Yamamoto Y, Takahashi T, Takahashi Y. Influence of Uridine Diphosphate Glucuronosyl transferase 2B7-161C>T Polymorphism on the Concentration of Valproic Acid in Pediatric Epilepsy Patients. Ther Drug Monit 2014; 36: 406-409.
- Ma Hongying, Zhang Ting, Gong Zhicheng, Zhou Boting, Zou Ming, Xiao Shan. Effect of UGT2B7 genetic variants on serum valproic acid concentration. J Central South University Medical Science 2013; 38: 766-772.
- Hans Carpay, William Arts. Outcome assessment in epilepsy: available rating scales for adults and methodological issues pertaining to the development of scales for childhood epilepsy. Epilepsy Res 1996; 24: 127-136.
- Hung CC, Ho JL, Chang WL, Tai JJ, Hsieh TJ, Hsieh YW. Association of genetic variants in six candidate genes with valproic acid therapy optimization. Pharmacogenomics 2011; 12: 1107-1117.
- Zhongliang Du, Yukun Jiao, Lianting Shi. Association of UGT2B7 and UGT1A4 Polymorphisms with Serum Concentration of Antiepileptic Drugs in Children. Med Sci Monit 2016; 22: 4107-4113.
- Zhongliang Du, Yukun Jiao, Lianting Shi. Association of UGT2B7 and UGT1A4 Polymorphisms with Serum Concentration of Antiepileptic Drugs in Children. Med Sci Monit 2016; 22: 4107-4113.
- Sun Yin-xiang, Zhuo Wen-yan, Lin Hong, Peng Zheng-ke, Wang Hua-ming, Huang Hao-wu. The influence of UGT2B7 genotype on valproic acid pharmacokinetics in Chinese epilepsy patients. Epilepsy Res 2015; 114: 78-80.