ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 5
Effect of sodium ferulate on IL-8 expression induced by Ox-LDL in human umbilical vein endothelial cells
1School of Public Health, Wuhan University, Wuhan, PR China
2Nanyang Medical College, Nanyang, PR China
3The First Affiliated Hospital of Nanyang Medical College, PR China
- *Corresponding Authors:
- Dongxian Zhang
Department of Basic of Basic Medical Sciences
Nanyang Medical College, P.R. China
E-mail: zdx990326@163.com - Yongyi Bi
School of Public Health
Wuhan University, PR China
E-mail: yongyibi@aliyun.com
Accepted date: January 02, 2016
In the present study we try to investigate the effect of Sodium Ferulate (SF) on Oxidized Low-Density Lipoprotein (Ox-LDL) induced IL-8 expression in Human Umbilical Vein Endothelial Cells (HUVEC). HUVECs were cultured in vitro and gene chip assay was used to analyse the regulation of IL-8 gene expression in HUVEC after the stimulation with Ox-LDL and SF+Ox-LDL, respectively. The IL-8 mRNA expression was measured by real-time PCR assay and protein expression by Western and ELISA assay. The results indicated that the expression of IL-8 gene was up-regulated by 6.1 fold after stimulation by Ox-LDL in HUVECs compared with that of the control. However, the expression of IL-8 gene was down-regulated by 3.3 fold after the stimulation by SF+Ox-LDL in HUVECs compared with that of in Ox-LDL treatment group. The real-time PCR results, Western and ELISA results showed that both the mRNA level and protein level of IL-8 increased significantly in Ox-LDL group compared with that of the control, and the expression decreased obviously in SF+Ox-LDL group compared with that of the Ox-LDL group. Take it together; Ox-LDL could induce the expression of IL-8, which could be inhibited by SF. Moreover, SF down-regulated the expression of IL-8, which could reduce the inflammatory response in HUVECs.
Keywords
Sodium ferulate, Human umbilical vein endothelial cell, Interleukin-8, Atherosclerosis
Abbreviations
AS: Atherosclerosis; h: Hours; HUVEC: Human Umbilical Vein Endothelial Cells; IL-8: Interleukin-8; mg: Milligram; ml: Millilitre; min: Minutes; mol/L: Molar Ox-LDL: Oxidized low-density lipoprotein; %: Percent; SF: Sodium Ferulate; VSMCs: Vascular Smooth Muscle Cells; VECs: Vascular endothelial cells.
Introduction
Pathological changes in the structure and function of Vascular Endothelial Cells (VECs) play an important role in Atherosclerosis (AS). Oxidized Low Density Lipoprotein (Ox- LDL) can cause secretion of surface adhesion molecules, pro-inflammatory cytokines and other inflammatory mediators by VECs, leading to vascular inflammation [1,2]. IL-8 acts as a regulator of immune and inflammatory response. It is secreted by endothelial cells, peripheral blood mononuclear cells and Vascular Smooth Muscle Cells (VSMCs), all of which are involved in the formation of AS [3]. IL-8 is believed to mediate the development of AS. Sodium Ferulate (SF) can reduce lipid peroxidation and clear free radicals from the body, so SF is now used to prevent and delay AS [4-7].
In order to investigate the influence of Ox-LDL on IL-8 expression of VECs and the protective effect of SF, we using a microarray assay to study the effect of SF on IL-8 expression induced by Ox-LDL and we try to investigate the biological mechanism.
Materials and Method
Cells and reagents
Gene microarray (CapitalBio Corporation, 22K Human Genome Array), HUVECs (CRL-1730, ATCC), DMEM medium (Hyclone), fetal bovine serum (Hangzhou Sijiqing Bioengineering Material Co., Ltd.), trypsin, penicillin-streptomycin, SF (Shanghai First Chemical Plant), Ox-LDL (Beijing Xiehe Sanyou Technology Development Co., Ltd.), Trizol (Invitrogen), GoldViewTMDNA dye (Shanghai SBS Genetech Co., Ltd.), LuxScan 3.0 software (CapitalBio Corporation), EvaGreen qPCR MassterMix II (CapitalBio Corporation).
Culture of HUVECs
Frozen HUVECs were resuscitated, placed in DMEM medium containing 10% fetal bovine serum and cultured at 37°C in a 5% CO2 humidified incubator. The culture medium was replaced every 2-3 days. After the monolayer of cells reached confluence, 0.25% trypsin was added to digest the cells. Logphase cells were harvested for further experiment.
Grouping
After reaching confluence, the culture medium was discarded and the cells were randomly divided into 3 groups: (1) control group: no drug treatment; (2) Ox-LDL group: 15 mg/mL Ox- LDL treatment for 24 h; (3) SF+OxLDL group: 5 μM SF treatment for 1 h, followed by 15 mg/mL Ox-LDL treatment for 24 h. The final volume was the same among the three groups.
Gene microarray
Total RNA extraction and identification: Total RNA extraction was performed by using Trizol reagent. RNA purity was assessed with an ultraviolet spectrophotometer, and its integrity by RNA gel electrophoresis using formaldehyde to keep RNA denatured. Fluorescence labelling of RNA sample: Using total RNA as initial template and T7 Oligo (dT) Primer (containing T7 promoter sequence), the synthesis of 1st-strand cDNA was started in the presence of CBC script. RNase H was used for the specific fragmentation of RNA, followed by addition of DNA polymerase and extension using RNA as primer. Then 2nd-strand cDNA was synthesized and double-stranded cDNA was purified. Using the synthesized cDNA as template, T7 Enzyme Mix was used to obtain cRNA, which was further purified using RNA Clean-up Kit. Reverse transcription was conducted using 2 μg cRNA, CBC Script II and random primer, and the products were purified with PCR NucleoSpin Extract II Kit. For the products of reverse transcription, Klenow fragment was used in fluorescence labelling reaction in the presence of random primer. During the extension of the complementary strands of DNA, Cy5-dCTP and Cy3-dCTP were added as the labelling reagents. The labelled products were purified with PCR NucleoSpin Extract II Kit and vacuum dried. Microarray hybridization and washing: Labelled DNA was dissolved in 80 μl hybridization buffer (3X SSC, 0.2% SDS, 5X Denhart’s, 25% formamide) and left at 42°C overnight. Hybridized DNA was first washed at 42°C in the buffer containing 0.2% SDS and 2X SSC for 5 min, then at room temperature in 0.2 × SSC for 5 min. The slide was dried and scanned. Microarray scan: Microarray images were extracted by LuxScan 3.0 software, which converted the images into digital signals. The data were normalized with Lowess method. Clustering analysis was conducted with Cluster3.0 software over the microarray data. CapitalBio Molecule Annotation System V 4.0 (MAS) was used to extract and analyse biological molecule relationships. Abnormal up-regulated and down-regulated genes were defined by the following threshold respectively: Ratio ≥ 2.0 and Ratio ≤ 0.5.
Western blot assay
Protein was extracted form threated cells. Separated the protein by using a 10% SDS-polyacrylamide gel, and transferred the separated protein on to a PVDF membrane. Blocked the transferred PVDF membrane with block buffer for 1 h and incubated the membrane with primary antibody (IL-8, β-actin) for 2 hours. Washing the PVDF membrane with TBST for 3 times and incubated the membranes with second antibody for 1 h. Washing the PVDF membrane with TBST for 3 times and the proteins were examined by using ECL chemiluminescence and exposed to x-ray film.
Real-time PCR assay
Using 1 μl of the synthesized cDNA as template, the PCR reaction system consisted of 2X Buffer 10 μl, 20X EvaGreen dye 0.6μl, Taq polymerase 0.3 μl (5 Unit), double-distilled water 6.9 μl, 10 μmol/L upstream and downstream primer 0.6 μl each. PCR procedures were as follows: predenaturation at 95°C for 5 min, denaturation at 95°C for 15 s, annealing at 50°C for 15 s, extension at 72°C for 20 s, fluorescence detection at 76°C for 3 s, for 40 cycles. Upstream primer of IL-8 was 5’-CATACTCCAAACCTTTCCACC-3’, and the downstream primer was 5’- AAACTTCTCCACAACCCTCTG-3’. The length of the amplified fragment was 112 bp. PCR products were analysed by 1.5% non-denaturing agarose gel electrophoresis. The hybridized microarray was scanned with a LuxScan 10 KA two-stand laser scanner.
ELISA assay
For ELISA detection, 100 μl of sample and standard was added, respectively, and left at 37°C for 90 min. Then 100 μl of biotinylated antibodies was added and left at 37°C for 60 min. The wells were washed with PBS for three times before reacting with 100 μl of ABC at 37°C for 30 min. The wells were further washed with PBS for five times and reacted with TMB at 37°C for 10-15 min. Stop solution was added and the OD value was measured with a microplate reader.
Statistical analysis
Results were reported as mean ± standard deviation. The mean values among groups were statistically compared by using ttest. p<0.05 was considered as a level of statistical significance.
Result
Microarray analysis
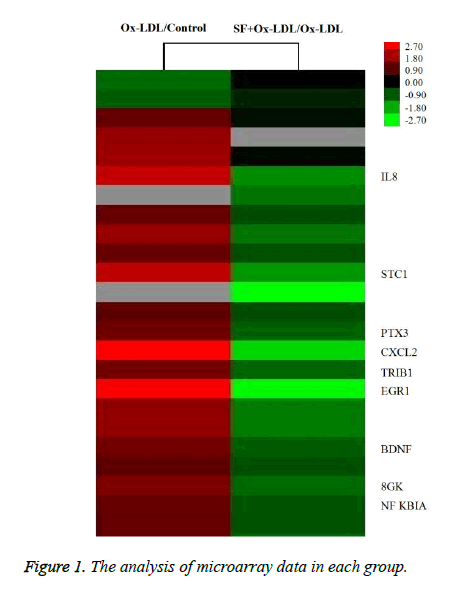
As indicated in Figure 1, the expression of IL-8 in HUVECs was up-regulated by about 4.2 fold after stimulation with Ox- LDL, while the IL-8 expression was down-regulated by about 2.8 fold in SF+Ox-LDL treatment compared with Ox-LDL treatment. Therefor SF could decrease the Ox-LDL induced IL-8 expression.
IL-8 mRNA expression by RT-PCR assay
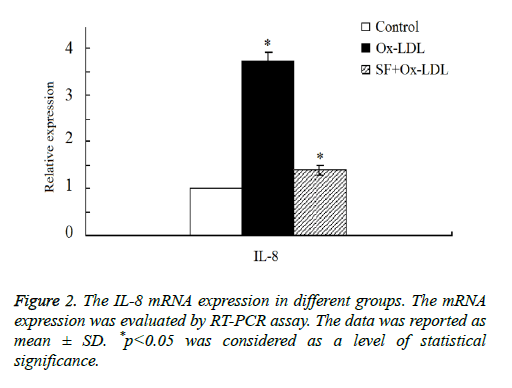
Then we double confirmed the expression of IL-8 expression by RT-PCR assay. As shown in Figure 2, the results indicated that there was a marked increase of IL-8 mRNA expression in Ox-LDL group compared with the control group. After the addition treatment of SF, IL-8 mRNA expression decreased significantly in SF+Ox-LDL group, and the IL-8 mRNA expression of the SF+Ox-LDL group was similar as it in the control group, which was consistent with the microarray analysis results. Therefore, Ox-LDL could induce the up-regulation of IL-8 mRNA, which could be inhibited by SF.
IL-8 protein expression
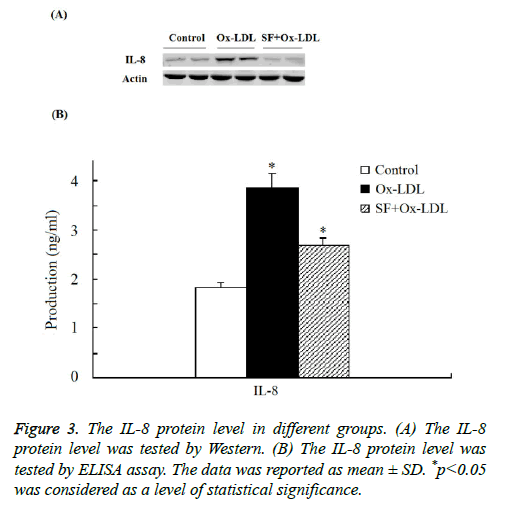
As indicated in Figure 3 by both Western and ELISA assay the expression of IL-8 protein was significantly increased in Ox- LDL group compared with the control group. And when treated with SF, the expression of IL-8 protein in the SF+Ox- LDL group decreased significantly to normal level compared to control group, which consistent with the microarray analysis and RT-PCR assay. Thus the Ox-LDL-induced IL-8 up-regulation could be inhibited by SF.
Discussion
AS is a common pathological condition for many cardiovascular diseases and mainly associated with endothelial dysfunction [8]. Abnormalities of VECs usually manifest as a weakening of anti-coagulation, anti-oxidant and anti-adhesion functions. Ox-LDL is one important reason behind VEC damage and the induced expressions of pro-inflammatory cytokines by VECs [9]. According to clinical trials, IL-8 is usually up-regulated for patients with coronary heart disease [10]. In animal experiments, IL-8 is also up-regulated in the plasma and the lesion sites [11].
IL-8 exerts a chemotactic effect on neutrophils and T-lymphocytes. IL-8 is similar to IL-1β, IL-6 and TNF-α in that it can activate the neutrophils and mediate inflammatory response and other pathological processes. It has been found that IL-8 promotes the adhesion of monocytes to endothelial cells and the invasion of the vascular wall, triggering the inflammatory response of the vascular wall. The adhesion between monocytes and endothelial cells further induces the activation of these two cells, leading to the secretion of IL-8 in the absence of other stimuli [12].
Studies showed that Ox-LDL significantly up-regulated IL-8 mRNA and protein expressions. However, the mechanism of anti-atherogenic effect of SF is generally unknown [13]. In the present study we found that SF inhibited the activity of Ox- LDL and down-regulated mRNA expression of IL-8, thereby reducing the secretion of IL-8 and the inflammatory response.
Acknowledgement
The work was supported by National Nature Science Foundation of China (U1404827) and Key Science and Technology Program of Henan Province.
References
- Cushing SD, Berliner JA, Valente AJ, Territo MC, Navab M, Parhami F, Gerrity R, Schwartz CJ, Fogelman AM. Minimally modified low density lipoprotein induces monocyte chemotactic protein 1 in human endothelial cells and smooth muscle cells. PNAS 1990; 87: 5134-5138.
- Chen YH, Lin SJ, Ku HH, Shiao MS, Lin FY. Salvianolic acid B attenuates VCAM-1 and ICAM-1 expression in TNF-alpha-treated human aortic endothelial cells. J Cell Biochem 2001; 82: 512-521.
- Inoue T, Komoda H, Nonaka M, Kameda M, Uchida T. Interleukin-8 as an independent predictor of long-term clinical outcome in patients with coronary artery disease. Int J Cardiol 2008; 124: 319-325.
- Wang B, Ouyang J, Liu Y, Yang J, Wei L. Sodium ferulate inhibits atherosclerogenesis in hyperlipidemia rabbits. J CardiovascPharmacol 2004; 43: 549-554.
- Li L, Zhang DX, Ruan L. Anatagonist effect of sodium ferulate on Ox-LDL. Chinese J Publ H 2010; 26: 689-691.
- Littlewood TD, Bennett MR. Apoptotic cell death in atherosclerosis. CurrOpinLipidol 2003; 14: 469-475.
- Liao JY. Protective effects of sodium ferulate in vascular endothelial function during cardiopulmonary bypass. Chin J ThoracCardiovascSurg 2011; 27: 221-223.
- Qiu YH. Functions and injury repair of vascular endothelial cells in artherosclerosis. J Clin Rehab Tissue Eng Res 2007; 11: 1927.
- Wang Y, Yin JH, Li HQ. Relationship between antibodies against oxLDL and occurrence of carotid atherosclerosis plaques. Chin J Publ H 2009; 25: 179-180.
- Liang LM, Huang ZH, Wei YS. Expressions of tumor necrosis factor and IL-8 in different types of coronary heart disease and the clinical significance. Chin J Lab Diag 2012; 16: 1180-1183.
- Wang Y, Wang HT, Zhang R. Intervention of IL-8 expression by rosuvastatin in rabbit model of artherosclerosis. J Jining Med Univ 2011; 34: 5-8.
- Azghani AO, Baker JW, Shetty S, Miller EJ, Bhat GJ. Pseudomonas aeruginosaelastase stimulates ERK signaling pathway and enhances IL-8 production by alveolar epithelial cells in culture. Inflamm Res 2002; 51: 506-510.
- Ou Yang Jing P, Wang BH, Liu YM, Yang JW, Wei L, Li K. Experimental study of the anti-atherogenesis effects of sodium ferulate in hyperlipidemia rabbits. Chin Pharmacol Bull 2002; 18: 207-210.