ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2015) Volume 26, Issue 4
Distribution of Hepatitis C Virus genotypes in patients with chronic Hepatitis C infection in Eastern Turkey
Türkan Toka Özer1, Mustafa Berktas2, Görkem Yaman3and Reha Erkoç4
1Department of Medical Microbiology, Mevlana University Faculty of Medicine, Konya, Turkey
2Department of Medical Microbiology, Lokman Hekim Hospital, Van, Turkey
3Department of Microbiology, Maltepe University Faculty of Medicine, Istanbul, Turkey.
4Department of Internal Medicine, Bezmialem Vakif University Faculty of Medicine, Istanbul, Turkey.
- Corresponding Author:
- Türkan Toka Özer
Department of Medical Microbiology
University of Mevlana
Faculty of Medicine
Yeni Istanbul
Cad. No: 235 42003
Selçuklu Konya/ Turkey
Accepted date: June 03 2015
Viral hepatitis is one of the most important liver diseases in the world. Duration of hepatitis C virus (HCV) infection and response to standard therapy is strongly related to HCV genotypes. Geographical distribution of HCV genotypes is also important for epidemiological studies. We aimed to determine the distribution of HCV genotypes and their prevalence in Eastern Turkey. Fifty eight Anti-HCV, LIA, HCV-RNA positive patients (35 male, 23 female) were included in this study. Genotypes were determined by reverse dot-blot hybridization method. Distribution of HCV genotypes in 58 patients were as follows: Genotype 1b in 53.4% (n=31), genotype 1a in 36.2% (n=21), genotype 1, 3, 4, 1a/1b in 1.7 % (n=1) for each. Genotype 1 was identified as the dominant type in 93.2% (n=54). HCV genotype could not be identified in 3.4% (n=2) patients. Genotype 1b was the most common HCV genotype in our region and this result is in accordance with the previous data of Turkey. Although genotype 1b is predominant, detection of genotype 1a in studies show that genotyping is beneficial for cost-effectiveness of chronic HCV infection. Thus, genotyping of HCV patients in our region guide clinicians in terms of monitoring and treatment success.
Keywords
Hepatitis C virus, Hepatitis C virus genotypes, Eastern Turkey, Chronic Hepatitis C, LIPA.
Introduction
Hepatitis C virus (HCV) is the most important factor of non-A, non-B hepatitis that may develop after transfusion and is seen sporadically in the community [1].
The prevalence of HCV infection in the world is estimated to be about 2.2-3%. Anti-HCV positivity in Turkey located in low endemic areas is between 0.5-2% [2]. Anti-HCV prevalence ranged between 1.2-2.6% in cohort studies conducted in healthy population, and such ratios have been reported as 0.05-1.5% among blood donors, 0.2-1% among health care workers, and 6.8- 51.6% among patients on hemodialysis [3].
HCV has a high viral heterogenecity. According to the nucleotide divergence, there are at least six genotypes, each of them containing a series of subtypes. Genotype frequencies vary by geographic region [4]. While genotype identification is important epidemiologically, it is also clinically useful in the assessment of response to therapy. Especially genotype 1b HCV-infected patients have poor response or no response to interferon. According to current treatment protocols, patients infected with genotype 2 or 3 respond to treatment faster and need lower doses of therapeutic agents [5, 6]. Thus, HCV genotyping is recommended before starting treatment.
There are no reports to show a well-attended HCV genotype distribution in our country, but small number of patient groups with regional studies are available. In this study, we aimed to determine distribution of HCV genotypes and their prevalence in Eastern Turkey.
Materials and methods
This study was conducted between March 2008 - May 2009 at Van Yüzüncü Yil University, Faculty of Medicine. A total of 126 patients diagnosed with chronic HCV infection were examined in this study. HCV-RNA was positive in only 58 of them and genotyping for HCV was performed on these patients only. Blood samples from the patients were obtained and assayed for Anti- HCV-positivity by MEIA method (AXSYM, Abbott, USA) and the test was confirmed with Line-Immune assay (INNO LIA HCV SCORE, Innogenetics, Belgium). The remaining serum samples were stored in Eppendorf tubes and stored at -80° C until PCR and genotyping analysis.
RNA extraction and quantitative determination of HCV-RNA
Sera stored at -80ºC was thawed at room temperature, vortexed and spun down by pulse-spin for 3-5 seconds. Exraction process was performed by MagNA Pure LC (Roche, USA). HCV-RNA quantification was performed with real time PCR technique using COBAS Taqman 48 (Roche, USA). Results were recorded as HCV RNA IU/ml.
HCV genotyping procedure
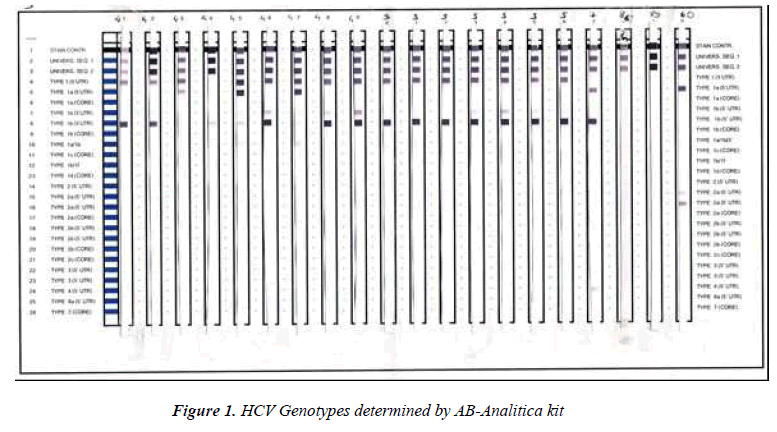
Genotyping was performed on the 58 patients after real time PCR analysis. HCV-RNA was extracted from stored serum samples using a QIAGEN kit (QIAamp MinElute Virus Spin Extraction kit, Germany). INNO LIPA HCV (Ampliquality HCV-TS; AB-Analitica, Italy) assay based on reverse dot-blot principle was used for HCV genotyping. The probes in this assay allow the identification of the following types: 1, 1a, 1b, 1a/b, 1c, 1d, 2, 2b, 2c, 3, 4, 5a, 6a, 7. Results were evaluated according to the schedule in kit insert. Results were confirmed by the manufacturer.
Of traits-descriptive statistics for continuous variables were referred to as Mean, Standard Deviation, Minimum and Maximum values, for categorical variables were expressed as number and percentage. Gender in terms of age, clinics and genotype in comparison of the means one-way Analysis of Variance (One-Way ANOVA) was performed. In determining the relationship between genotype, gender and clinics, Chi-square test and multiple correspondence analysis was performed. In the calculations, the level of statistical significance was taken as 5%, and SPSS statistical software was used in the calculations.
Ethical approval was granted by the Ethical Committee of Yüzüncü Yil University (No:2008/04). Informed consent was obtained from all patients taking part in the study.
Results
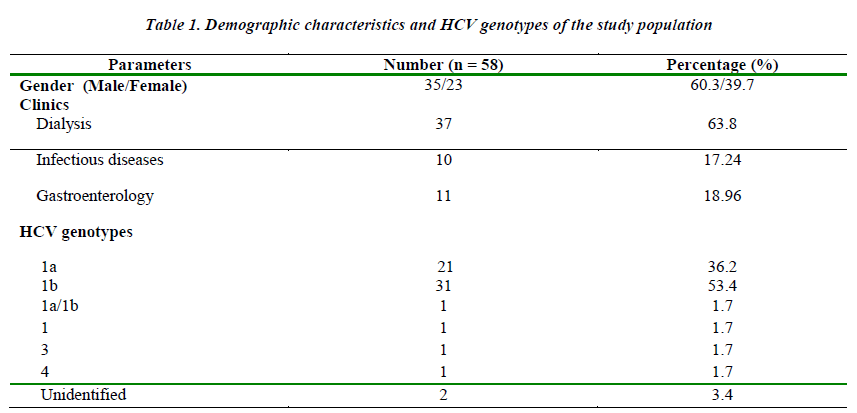
Distribution of genotypes and clinics were shown on Table 1. In 2 of the 58 patients (3.4%) genotyping procedures failed and no band appeared. Genotype 1 was identified as the dominant type in total of 54 patients (93.2 %) (Table 1). Genotypes of patients assessed considering the mean age by gender; HCV patients, 35 (60.3%) were male (mean age 50.43 ± 13.8), 23 (39.7%) were female (mean age 54.70 ± 15.7). The mean age of male and the female participants were similar. There was no significant difference among clinics of the patients for HCV genotypes. According to gender; in genotype 1b patients, 21 (67.7%) were male and 10 (32.3%) were female; in genotype 1a patients, 10 were male (47.6%) and 11 were female (52.4%). There was no significant difference due to gender of the patients for HCV genotypes. The mean age of patients with genotype 1b was 51.19 ± 15.46, and the mean age of patients with genotype 1a was 53.48 ± 14.25, respectively. There was no significant difference between ages of the patients and HCV genotypes. Although not statistically significant; the cases of patients between the ages of 40-60 years were more common from the dialysis unit, and the most common genotype for this population was genotype 1b. However, the cases between the ages of 20-40 years were mostly from the other clinics and the most common genotype was genotype 1a.
Discussion
HCV infection is still one of the major health problems in the world [7]. HCV nucleotide sequences, which are different from each other up to 30 % of the time, are divided into six genotypes and more than 80 subtypes [8]. While some HCV genotypes are distributed all over the world, some genotypes are limited to certain geographic areas. HCV type 1 infections are common worldwide. Genotypes 1, 2 and 3 are more common genotypes and are observed mostly at Europe, North America, China, Japan and Australia. There are more significant differences in the distribution of subtypes. Type 1a often was found in Northern Europe and North America, and Iran; type 1b is the most common genotype in Japan, South and Eastern Europe. Genotype 1 was found in 71 % of the infected population in USA and type 2 is rarer than type 1 in the world. HCV subtypes 2a and 2b are relatively common in North America, Europe and Japan and subtype 2c is most common in northern Italy [9-12].
The majority of patients are infected with HCV genotype 3 in Thailand, Malaysia, India and Pakistan [13]. HCV genotype 4 appears to be prevalent in North Africa and the Middle East [9, 14-16]. Genotype 4 infection is reported to be dominant in Yemen, Kuwait, Saudi Arabia, Iraq, Zaire, Gabon and Gambia. The HCV genotype (type 1b was dominant) distribution in Algeria is different from those in other northern countries of Africa [1, 12]. Genotypes 5 and 6 seem to be confined to South Africa and Southeast Asia, respectively [9, 14-16]. In Hong Kong, genotype 6a is the the most common type of infection [17]. Genotype 6 is distributed primarily in the southern region of China [18].
In studies conducted in Turkey on HCV genotypes, genotype 1 HCV infection was found to be about 90%, the majority of them are type 1b, type 2, 3 and 4 are found less frequently in HCV infections [19-22]. Genotype 1a is reported as higher in Izmir, Ankara, and Diyarbakir (19.1- 33.3%) [19, 23].
In our study, the detection of genotype 1 in the 54's (93.2%) of 58 chronic HCV- infected patients genotype shows that genotype 1 is prevalent in our region. These results are compatible with previous reports from our country. In the evaluation of our work, subtypes of genotype 1b were detected in 53.4% of 31 patients. This result is consistent with other studies, in that type 1b appears to be dominant. In our study, the most common types of HCV 1b genotype determination is a finding consistent with the literature of our country and the world. However, in 21 patients with genotype 1a in the second row is determined as 36.2%. This region shares a border with Iran and the prevalence of genotype 1a in Iran may be responsible for this. Many tourists from Turkey travel to this region from Iran. Kinship with the people of Iran is also a consideration. Genotype 1a is the most common genotype in Iran [10]. Ethnic and population movements in the region also may change the relative frequency of genotypes [24-25].
The results clearly show that there is no variation among HCV genotypes and gender as the different HCV genotypes were distributed with the same ratio in males and females in this study. In Libya, the prevalence of HCV genotype 1 was found to be mostly in males, while genotype 4 has been found more frequently in females [12]. Both genotype 3 (81%) and 1 (80%) were more frequent in males [13].
Although not statistically significant, the cases between the ages of 40-60 years were more common from dialysis unit, and the most common genotype for this population was genotype 1b. However, the cases between the ages of 20-40 years were mostly from the other clinics and the common genotype was genotype 1a. Studies showed that patients infected with genotype 1b or 2 were older than patients infected with genotype 1a, 3 and 4 [13]. Rouabhia et al. reported that genotype 1 is associated with the age group younger than 60 and is lower in older age group. To the contrary, the HCV non-genotype 1 was higher among the patients over 60 years [12]. In studies in Turkey in hemodialysis patients, genotype 1b was found to be the dominant one [26, 27].
HCV genotyping is an epidemiological marker for prognosis of treatment response in patients. Therefore, many efforts around the world have been devoted to HCV genotyping and detection. For achieving the mentioned goal, a simple, sensitive and reliable system is needed. HCV genotyping is performed by several different methods. Although sequence analysis is the gold standard for identification of HCV genotypes, it is impractical for routine clinical laboratories. Today, the most widely used means of detection of HCV genotypes is the reverse dotblot hybridization method, which is based on the principle of 'line probe assay’(LIPA). LIPA permits rapid determination of the types and subtypes of HCV, and might aid detection of new HCV genotypes [28, 29].
High costs of drugs for the treatment of HCV, which has quite a lot of side effects, enhanced the importance of genotype determination. However, depending on the genotype, the different responses to interferon therapy is not known precisely [30]. HCV infection is a progressive disease and to have the chance to get rid of the virus with treatment and to prevent HCC in patients responding to treatment requires early initiation of therapy in our country. Thus, population-based epidemiological studies about HCV has critical importance in our region and in our country.
As a result of this study; genotypes of patients with chronic HCV infection were determined as 53.4% genotype 1b, 36.2% genotype 1a, 1.7% genotype 1, genotype 3, genotype 4 and genotype 1a/1b. Genotype 1 (93.2%) was the most common HCV genotype in our region. In studies conducted in our country although the predominant type is genotype 1b, the determination of genotype 1a at a rate of 36.2% reveals the necessity of genotype determination in the effective treatment of chronic HCV infection. The results of this study will guide clinicians in successful monitoring and treatment of HCV patients in our region.
Conflict of interests
None declared.
Acknowledgments
This study was financially supported by the Yüzüncü Yýl University Department of Scientific Research Project as No. 2008-TFI-047 project. We would like to thank Prof. Dr. Siddik Keskin and Dr. Abdurrahman Özer, who lent their support to us in the study. This study was presented as oral presentation at 5th EACID 15-18 May 2013, Tirana, Albania.
References
- Abacioglu H. Hepatit C Virüsü: In: TemelveKlinikMikrobiyoloji. GünesKitabevi, Ankara 1999; pp881-889.
- Köksal Y, Leblebicioglu H. KronikHepatitlerinTaniveTedavisindeGüncelYaklasimlar. BilimselTip Yayinevi, Ankara 2009; pp 11-25.
- Tahan V, Ozdogan O, Tozun N. Epidemiology of viral hepatitis in the Mediterranean basin. RoczAkad Med Bialymst 2003; 48: 11-17.
- Simmonds P, Bukh J, Combet C. Deléage G, Enomoto N, Feinstone S, Halfon P, Inchauspé G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E,Shin-I T, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. Consensus Proposals for a Unified System of Nomenclature of Hepatitis C Virus Genotypes. Hepatology 2005; 42: 962-973.
- Cristina J, delPilar Moreno M, Moratorio G. Hepatitis C virus genetic variability in patients undergoing antiviral therapy. Virus Research 2007; 127: 185-194.
- Ahmed F, Jacobson IM. Treatment of Relapsersafter Combination Therapy for Chronic Hepatitis C. Infect Dis Clin N Am 2006; 20: 137-153.
- Lee CM, Hung CH, Lu SN, Changchien CS. Hepatitis C Virus Genotypes. Clinical Relevance and Therapeutic Implications. Chang Gung Med J 2008; 31(1): 16-25.
- Scott JD, Gretch DR. Hepatit C ve Gvirüsleri: In: KlinikMikrobiyoloji. Cilt 2. 9. Baski Atlas Kitapçilik, Ankara 2009; pp 1437-1452.
- Zein NN. Clinical significance of Hepatitis C Genotypes.ClinMicrobiol Rev 2000; 13(2): 223- 235.
- Kabir A, Alavian SM, Keyvani H. Distribution of hepatitis C virus genotypes in patients infected by different sources and its correlation with clinical and virological parameters: A preliminary study. Comp Hepatol 2006; 5: 4.
- Magiorkinis G, Magiorkinis E, Paraskevis D, Ho SY, Shapiro B, Pybus OG, Allain JP, Hatzakis A. The Global Spread of Hepatitis C Virus 1a and 1b:APhylodynamic and Phylogeographic Analysis. PLoS Med 2009; 6(12):1-12.
- Rouabhia S, Sadelaoud M, Chaabna-Mokrane K, Toumi W,Abenavoli L. Hepatitis C virus genotypes in north eastern Algeria: A retrospective study. World J Hepatol 2013; 5(7): 393-397.
- Mohamed NA, Rashid ZZ, Wong KK, Abdullah SA, Rahman MM. Hepatitis C genotype &associated risks factors of patients at University Kebangsaan Malaysia Medical Centre. Pak J Med Sci 2013; 29(5):1142-1146.
- Ayesh BM, Zourob SS, Abu-Jadallah SY, Shemer- Avni Y. Most common genotypes and risk factors for HCV in Gaza strip: a cross sectional study. Virol J 2009; 6: 105.
- Yu ML, Chuang WL. Treatment of chronic hepatitis C in Asia: WhenEast meets West. J Gastroenterol andHepatol 2009; 336-345.
- Shemis MA, El-Abd DM, Ramadan DI, El-SayedMI, Guirgis BS, Saber MA, Azzazy HM. Evaluation of Multiplex Nested Polymerase Chain Reaction for Routine Hepatitis C Virus Genotyping in Egyptian Patients. Hepat Mon 2012; 12(4): 265- 270.
- Nausbaurn JB. Genomic subtypes of Hepatitis C virüs:epidemiology, Diagnosis and clinical consequences.Bull SocPatholExot. 1998; 91(1):29-33.
- Akkarathamrongsin S, Praianantathavorn K,Hacharoen N, Theamboonlers A, Tangkijvanich P,Tanaka Y, Mizokami M, Poovorawan Y. Geographic Distribution of Hepatitis C Virus Genotype 6 Subtypes in Thailand. J Med Virol2010; 82(2): 257-262.
- Buruk CK, Bayramoglu G, Reis A, Kaklikkaya N, TosunI, AydinF. Determination of Hepatitis C Virus Genotypes Among Hepatitis C Patients in Eastern Black Sea Region, Turkey. MikrobiyolBul2013; 47(4): 650-657.
- Bozdayi G, Rota S, Verdi H, Derici U, Sindel S, Bali M, Basay T. The presence of hepatitis C virus (HCV) infection in hemodialysis patients and determination of HCV genotype distribution.MikrobiyolBul 2002; 36: 291-300.
- Tezcan S, Ulger M, Aslan G, YarasS, AltintasE, Sezgin O, EmekdasG, Gürer-Giray B, Sungur MA. Determination of Hepatitis C Virus Genotype Distribution in Mersin Province, Turkey.MikrobiyolBul 2013; 47(2): 332-338.
- Türkoglu S, Bozaci M, Çakaloglu Y. Ikincikusak“core genotiplemesi”ilehepatit C virus genotiplerininarastirilmasi, 3. UlusalHepatolojiKongresi, Istanbul, May, 41, 1999.
- Erensoy S, Göksel S, Akarca US, Özkahya M, Canatan D. Genotyping of Hepatitis C Virus by Direct Sequence Analysis of Polymerase Chain Reaction Products. Flora 2002; 7: 104-111.
- Akhan S. Hepatit C Virüsü. In: EnfeksiyonhastaliklariveMikrobiyolojisi: Cilt 2. 3. Baski. Nobel Tip Kitabevleri, Antalya 2008; pp 1911- 1928.
- Durmaz R. HCV mutasyonlari. In: Viral Hepatit2005.VHSD 2005; pp 170-175.
- Selçuk H, Kanbay M, Korkmaz M, Gur G, AkcayA, Arslan H, Ozdemir N, Yilmaz U, Boyacioglu S. Distribution of HCV Genotypes in Patients with End-Stage Renal Disease According to Type of Dialysis Treatment. Dig Dis Sci 2006; 51: 1420–1425.
- Bozdayi AM, Aslan N, Bozdayi G, Türkyilmaz AR, Sengezer T, Wend U, Erkan O, Aydemir F, Zakirhodjaev S, Orucov S, Bozkaya H, Gerlich W, Karayalçin S, Yurdaydin C, Uzunalimoglu O. Molecular epidemiology of hepatitis B, C and Dviruses in Turkish patients. Arch Virol 2004; 149: 2115-2129.
- Majidzadeh-A K, Morovvati A, Soleimani M, Langeroudi AG, Merat S, Jabbari H. Development and Application of an In-house Line Probe Assay for Hepatitis C Virus Genotyping. Hepat Mon 2013;13(5): e6767.
- Sanlidag T, Akçali S, Ozbakkaloglu B, Ertekin D, Akduman E. Distribution of hepatitis C virus genotypes in Manisa region, Turkey. MikrobiyolBul 2009; 43(4): 613-618.
- Hofmann WP, Zeuzem S, Sarrazin C. Hepatitis C virus-related resistance mechanisms to interferon alpha-based antiviral therapy. J ClinVirol 2005; 32: 86-91.