ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 13
Detection and molecular identification of human Giardia isolates in the West of Iran
1School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
2Department of Parasitology and Mycology, School of Medicine, Kurdistan University of Medical Sciences, Kurdistan, Iran
- *Corresponding Author:
- Ali Haghighi
School of Medicine
Shahid Beheshti University of Medical Sciences, Iran
Accepted May 09, 2017
Giardia lamblia, one of the most common intestinal protozoan pathogens worldwide, causes human gastrointestinal diseases. Because of its high prevalence and major impact on the patients’ quality of life, giardiasis has become one of the most important public health problems. We aimed to evaluate the prevalence and assemblages of G. lamblia isolated from referred individuals to the medical laboratories in the center of Kurdistan province, West of Iran. Direct wet mount and formalin-ether sedimentation technique were applied for microscopy determination of Giardia infection in 1383 obtained stool samples. DNA was extracted from positive stool samples and amplified by Nested-PCR using specific primers of G. lamblia Trios phosphate isomerase (TPI) gene. Genotyping and assemblages was determined using sequence analysis of the TPI gene. Based on microscopic examination, out of the 1383 samples, 23 (1.66%) G. lamblia were detected. Genotyping data indicated 12 (52%) cases as assemblage a cluster, and 11 (48%) cases as assemblage B cluster. Molecular analysis in this study reveal that assemblages A and B are the most common types of human giardiasis in Kurdistan province and giardiasis could be a zoonosis disease. Molecular identification of Giardia isolates from animals and control programs is recommended to prevent the spreading of giardiasis in humans.
Keywords
Giardia lamblia, Molecular identification, Genotyping, Triose phosphate isomerase, Zoonose, Kurdistan province, Iran.
Introduction
Historically, Giardia lamblia was a very common causative agent of diarrhea [1]. This organism was first observed by van Leeuwenhoek in 1681 while he checked his own stools under the microscope [2]. About 280 million are currently estimated to be infected with Giardia each year throughout the world [3]. Because of its high prevalence and major impact on the patients’ quality of life, giardiasis has become one of the most important public health problems [4,5]. This neglected disease [6], can be debilitating for healthy individuals and can result in significant morbidity, especially in children [7]. Transmission of G. lamblia generally occurs by fecal-oral transmission through food, drinking water, or recreational water contaminated with feces containing the cysts. There have also been some waterborne outbreaks from water contaminated with feces [1].
G. lamblia complex is found in the intestine and can infect a wide variety of vertebrate hosts. This complex is morphologically similar, but with a genetically different assemblage [8]. Some assemblages of G. lamblia infect a wide range of mammal hosts, whereas others appear to be restricted to groups such as cats or dogs, and some are known to infect only one host species. These assemblages include A, B, C, D, E, F, G, and H. Assemblages A and B isolates are known to infect a broad range of hosts, including humans, livestock, cats, dogs, beavers and guinea pigs. Assemblages C and D infect dogs and cats, while assemblage E infects cattle, sheep, and goats. Furthermore, assemblage F infects only cats, assemblage G infects rats, and assemblage H infects seals. Therefore, only assemblages A and B can infect human and other mammals. While the other assemblages have so far not been detected in humans [9].
Most targeted genes used for characterizing G. lamblia species rely on the analysis of the small subunit ribosomal DNA (SSUrDNA), the Elongation Factor 1 Alpha (ef1a), the triosephosphate isomerase (TPI), and the glutamate dehydrogenase (GDH) genes [10]. Giardiasis is a major problem in Iran [11], as well as in Kurdistan province, west of Iran. There is however, no data currently available concerning the prevalence of different Giardia genotypes in this region. We aimed to assess the prevalence of G. lamblia genotypes (assemblages) in center of Kurdistan province.
Materials and Methods
In this study, 1383 fecal samples from 1 June 2015 to 30 November 2016 were collected from individuals who had been referred to 14 medical laboratories in center of Kurdistan province. Giardia cyst or trophozoite were identified in samples using direct wet mount and formalin-ether sedimentation technique by microscopic observation (Zeiss, Germany, 40X magnification). The isolates of positive samples were preserved in alcohol and kept at 4°C until used for DNA extraction [12].
DNA extraction
Approximately 300 μl of fecal suspension was washed three times with distilled water to remove the trace of alcohol and then genomic DNA was extracted directly from feces using the DNA extraction kit according to the manufacturer’s instructions (YTA, FavorGen, Cat. No YT9032, Taiwan) with the addition of glass milk matrix method and freeze-thaw cycles (10 min). Finally the suspended DNA samples in lyses buffer were frozen in liquid nitrogen and thawed at 90°C in water bath. The extracted DNA was stored in sterilized tubes and frizzed at -20°C until tested for PCR [13].
Primers and PCR amplification
To identify the assemblages of G. lamblia, nested PCR based on the genetic loci of TPI was used to amplify a fragment (530-605 bp) of the TPI gene using two sets of oligonucleotide primers. A fragment of 605bp was amplified using primers AL3543 [5’-AAATIATGCCTGCTCGTCG-3’] and AL3546 [5’-CAAACCTTITCCGCAAACC-3’] for primary PCR and a fragment of 530 bp was amplified using primer AL3544 [5’- CCCTTCATCGGIGGTAACTT-3’] and AL3545 [5’- GTGGCCACCACICCCGTGCC-3’] for secondary PCR, as originally described by Sulaiman et al. [14]. Primers contained degenerate bases to enable amplification of isolates across all assemblages. The primer sequences were checked for specificity by conducting BLAST searches of the NCBI GenBank database. The primers were synthesized by Bioneer® (Corp., Chungwon, Korea).
A PCR kit (Ampliqon ApS, literbuen 11, DK- 2740 Skovlunde, Denmark) was used to accelerate and increase the quality of PCR products. Nested TPI PCR was performed according to Sulaiman and colleagues’ method [14]. Primary amplification was carried out in 35 cycles (94°C for 45 s, 58°C for 45 s, and 72°C for 1 min) with an initial denaturation (94°C for 5 min) and a final extension (72°C for 5 min). For secondary amplification, 35 cycles (94°C for 45 s, 58°C for 45 s, and 72°C for 1 min) were used, with identical initial denaturation and final extension conditions. Both positive and negative controls were included in each round of PCR to validate results. Reactions were visualized in UV on 1.5% agarose gels stained with ethidium bromide.
Sequence analysis of TPI gene
Genotyping was performed using sequence analysis on the 23 PCR products of G. lamblia. The secondary PCR products were sequenced using AL3544 primer. Nucleotide sequences were edited manually and the sequence representatives for each of the assemblages identified in the studies were submitted to the GenBank/EMBL/DDBJ database under accession No. of KY359204 to KY359226 (Table 3).
Phylogenetic analysis
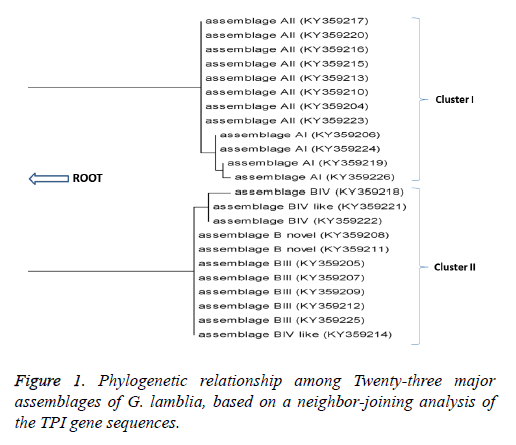
Sequence information was obtained for a representative isolate of each of the Assemblages A and B from the NCBI database. The sequences were aligned using ClustalW2. The resulting sequences were analyzed and compared with similar G. lamblia sequences deposited in GenBank using the Basic Local Alignment Search Tool (BLAST) program. Based on TPI sequences, a dendrogram was constructed using the neighborjoining (NJ) method in MEGA5 software and bootstrap analysis with 1,000 resamplings (Figure 1).
Results
Microscopy detection
Of the 1383 stool samples using parasitology methods, the overall frequency of the intestinal parasitic infections was 21.5%. Based on microscopic examination the prevalence of G. lamblia infection was found to be 1.66% (23/1383). Table 1 shows some of the patients’ demographic data such as sex, age, and residential area base on being symptomatic or not. Eleven (48%) infected Giardia patients with gastrointestinal symptoms were children under 12 years of age and 7 (30%) were older).
| Variable | Clinical features | ||
|---|---|---|---|
| Symptomatic | Asymptomatic | ||
| Sex | Male | 9 | 4 |
| Female | 9 | 1 | |
| Age group (years) | <6 | 3 | 0 |
| 06-Dec | 8 | 0 | |
| Dec-18 | 1 | 1 | |
| 18-30 | 4 | 2 | |
| 30-50 | 1 | 1 | |
| >50 | 1 | 1 | |
| Location | Urban | 17 | 3 |
| Rural | 1 | 2 | |
Table 1. Distribution of Giardia lamblia based on sex, age and location of the referred patients in Kurdistan province, Iran.
All of the patients who lived in rural areas had close contact with animals. Also, most infected patients were from urban areas and did not have any contact with animals and were seen in summer and autumn (Table 2).
| Variable | Category | Frequency | Assemblages | |
|---|---|---|---|---|
| A | B | |||
| Sex | Male | 13 | 8 | 5 |
| Female | 10 | 4 | 6 | |
| Age group (years) | <6 | 3 | 0 | 3 |
| 6-12 | 8 | 3 | 5 | |
| 12-18 | 2 | 0 | 2 | |
| 18-30 | 6 | 6 | 0 | |
| 30-50 | 2 | 2 | 0 | |
| >50 | 2 | 1 | 1 | |
| Source of drinking water | Treated | 21 | 10 | 11 |
| Untreated | 2 | 2 | 0 | |
| Contact with domestic animals | No | 20 | 10 | 10 |
| Yes | 3 | 2 | 1 | |
| Location | Urban | 20 | 11 | 9 |
| Rural | 3 | 1 | 2 | |
| Seasons | Spring | 0 | 0 | 0 |
| Summer | 16 | 7 | 9 | |
| Autumn | 7 | 5 | 2 | |
| Winter | 0 | 0 | 0 | |
| Clinical features | Symptomatic | 18 | 8 | 10 |
| Asymptomatic | 5 | 4 | 1 | |
Table 2. Frequency of Giardia lamblia genotypes in patients with giardiasis cases based on demographic characteristics.
PCR and sequencing
The DNAs were successfully extracted from all positive samples and the PCR fragments were successfully visualized on gel electrophoresis. The targeted TPI sequence fragment was sequenced in all isolates. After sequencing the forward strand of the positive PCR, their assemblages were determined using the standard nucleotide BLAST algorithm provided by NCBI (http://www.ncbi.nlm.nih.gov/). The genotyping results indicated 12 (52%) isolates as assemblage A (AI and AII) and 11 (48%) as assemblage B (BIII, BIV and B novel) (Table 3).
| Sample ID | Assemblage | Accession No. |
|---|---|---|
| GBS-IR71 | AII | KY359204 |
| GBS-IR99 | BIII | KY359205 |
| GBS-IR147 | AI | KY359206 |
| GBS-IR179 | BIII | KY359207 |
| GBS-IR185 | B novel | KY359208 |
| GBS-IR244 | BIII | KY359209 |
| GBS-IR334 | AII | KY359210 |
| GBS-IR446 | B novel | KY359211 |
| GBS-IR593 | BIII | KY359212 |
| GBS-IR669 | AII | KY359213 |
| GBS-IR704 | BIV like | KY359214 |
| GBS-IR725 | AII | KY359215 |
| GBS-IR783 | AII | KY359216 |
| GBS-IR933 | AII | KY359217 |
| GBS-IR1048 | BIV | KY359218 |
| GBS-IR1147 | AI | KY359219 |
| GBS-IR1159 | AII | KY359220 |
| GBS-IR1175 | BIV like | KY359221 |
| GBS-IR1178 | BIV | KY359222 |
| GBS-IR1213 | AII | KY359223 |
| GBS-IR1290 | AI | KY359224 |
| GBS-IR1370 | BIII | KY359225 |
| GBS-IR1374 | AI | KY359226 |
Table 3. Characterization of Giardia lamblia assemblages detected in humans in Kurdistan, based on sequence analysis.
Phylogenic analysis
Phylogenetic analysis (Figure 1) grouped the isolates into two main clusters namely cluster I which contained assemblage A, with most of the isolates clustering within this clade with sub- assemblages AI and AII (Figure 1). And Cluster II, contained assemblage B with sub-assemblages BIII, BIV and B novel.
Discussion
G. lamblia has been associated with morbidity in animals and humans. It is also recognized as a human foodborne or waterborne pathogen and zoonosis disease [15]. G. lamblia has two major genotypes, which are referred to as “Polish” and “Belgian”, or assemblage A and B, respectively [16]. Current molecular data indicates that G. lamblia is a complex genus grouped into eight main assemblages (A-H ) [9]. So far assemblages C-H have not been detected in humans [17]. Moreover, it has been suggested that the G. lamblia assemblages differ from each other with respect to pathogenicity and clinical presentations [8]. For this reason, using molecular techniques is necessary to determine the assemblages infecting humans and those leading to clinical symptoms. Based on microscopic examination, we found that the prevalence of giardiasis infection was 1.66% (23/1383). This prevalence is close similar to reports from Qazvin [18] and Karaj [19] with a prevalence of 0.85% and 3.8%, respectively. However, high prevalence rates were reported in Kirkuk (10.31%) [20], Shiraz (10.6%) [11], Shahre-Ray (25.8%) [21], and Hamadan (20%) [22].
We identified both assemblages A and B, and an approximately equal distribution between these two in the study population, similar to distributions found Tabriz [23,24], Ahvaz [25] and Saudi Arabia [26]. In contrast, studies in Australia [27], Egypt [28] India [29], Kuala Lumpur [30], UK [31] and Bangladesh [32] showed that infections with assemblage B were more prevalent than infection with assemblage A, whereas this differed from findings in Tehran [13], Fars province [33], and Isfahan [34], where more infections were associated with assemblage A, and only few with assemblage B. The question, whether giardiasis is a zoonosis defined as an animal related disease that is transmissible to humans has been raised in the past [35]. For over twenty years, the World Health Organization has considered Giardia to have zoonotic potential [36]. Moreover, evidence for waterborne or anthroponiotic routes of transmission is still scarce [37]. Recent evidence shows that G. lamblia assemblage A and B are possibly zoonotic. In other words, sub-assemblage AI, AII, BIII, and BIV are potentially zoonotic, whereas sub-assemblage AIII is found exclusively in animals [38]. The predominance of zoonotic giardiasis in humans in Kurdistan is consistent with studies from some developed and developing countries such as New Zealand [39], Eastern England [40] and India [29]. In contrast, the predominance of anthroponiotic giardiasis in humans has been reported from Southern Iran [33], Tehran [13], Shiraz [11], Yemen [41], and Malaysia [30].
The findings showed that 66.6% of assemblage A and 90% of assemblage B were symptomatic. We found no significant association between assemblage and clinical symptoms consistent with findings from Ahvaz [25], Southern Iran [33], and Northeast China [42]. With respect to the relationship between assemblage and clinical symptoms, some studies show that assemblage B was associated with clinical symptoms and some studies show that clinical symptoms related to assemblage A. However, some other studies show no relationship between type of assemblages and clinical symptoms. In two studies in Kuala Lumpur [30] and Saudi Arabia [26], clinical symptoms were strongly associated with assemblage B; while in studies from Isfahan [34], Bangladesh [32], and Egypt [43], assemblage A were more likely to be symptomatic compared with assemblage B. In a study from Southern Iran [33], assemblage AII was more frequently associated with abdominal pain, nausea and vomiting.
Conclusion
Molecular analysis in this study revealed that assemblages A and B are the most common types of human giardiasis in Kurdistan province and giardiasis could be a zoonosis. Molecular identification of Giardia isolates from animals and control programs is recommended to prevent the spreading of giardiasis in humans.
Acknowledgements
We thank all technicians and personnel working in the medical laboratories, School of Medicine, Kurdistan University of Medical Sciences and also those who work in the Department of Medical Parasitology and Mycology, School of Medicine, Shahid Beheshti University of Medical Sciences who contributed to the study.
Financial and Scientific Support
This work was part of the PhD thesis of Fares Bahrami under supervision of Prof. Ali Haghighi and Dr. Ghasem Zamini, which was supported financially by Kurdistan University of Medical Sciences (Grant No. 335) and Shahid Beheshti University of Medical Sciences (Grant No. 6598).
References
- Adam EA, Yoder JS, Gould LH, Hlavsa MC, Gargano JW. Giardiasis outbreaks in the United States, 1971-2011. Epidemiol Infect 2016; 144: 2790-2801.
- Ortega YR, Adam RD. Giardia: overview and update. Clin Infect Dis 1997; 25: 545-549.
- Ankarklev J, Jerlström-Hultqvist J, Ringqvist E, Troell K, Svärd SG. Behind the smile: cell biology and disease mechanisms of Giardia species. Nat Rev Microbiol 2010; 8: 413-422.
- Ajjampur S, Koshy B, Venkataramani M, Sarkar R, Joseph A, Jacob K, Ward H, Kang G. Effect of cryptosporidial and giardial diarrhoea on social maturity, intelligence and physical growth in children in a semi-urban slum in south india. Ann Trop Paediatr 2011; 31: 205-212.
- Simsek Z, Zeyrek FY, Kurcer MA. Effect of Giardia infection on growth and psychomotor development of children aged 0-5 years. J Trop Pediatr 2004; 50: 90-93.
- Savioli L, Smith H, Thompson A. Giardia and Cryptosporidium join the 'Neglected Diseases Initiative'. Trends Parasitol 2006; 22: 203-208.
- Escobedo AA, Alvarez G, González ME, Almirall P, Cañete R. The treatment of giardiasis in children: single-dose tinidazole compared with 3 days of nitazoxanide. Ann Trop Med Parasitol 2008; 102: 199-207.
- Cacciò SM, Ryan U. Molecular epidemiology of giardiasis. Mol Biochem Parasitol 2008; 160: 75-80.
- Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev 2011; 24: 110-140.
- Monis PT, Andrews RH, Mayrhofer G, Ey PL. Molecular systematics of the parasitic protozoan Giardia intestinalis. Mol Biol Evol 1999; 16: 1135-1144.
- Rayani M, Unyah NZ, Hatam G. Molecular identification of giardia duodenalis isolates from fars province, iran. Iran J Parasitol 2014; 9: 70.
- Sousa MC, Morais JB, Machado JE, Poiares-da-Silva J. Genotyping of Giardia lamblia human isolates from Portugal by PCR-RFLP and sequencing. J Eukaryot Microbiol 2006; 53: S174-176.
- Babaei Z, Oormazdi H, Akhlaghi L, Rezaie S, Razmjou E, Soltani-Arabshahi S, Meamar A, Hadighi R. Molecular characterization of the iranian isolates of giardia lamblia: Application of the glutamate dehydrogenase gene. Iran J Public Health 2008; 37: 75-82.
- Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, Das P, Lal AA, Xiao L. Triosephosphate isomerase gene characterization and potential zoonotic transmission of giardia duodenalis. Emerg Infect Dis 2003; 9: 1444-1452.
- Yoder JS, Gargano JW, Wallace RM, Beach MJ; Centers for Disease Control and Prevention (CDC). Giardiasis surveillance-United States, 2009-2010. MMWR Surveill Summ 2012; 61: 13-23.
- van Keulen H, Macechko PT, Wade S, Schaaf S, Wallis PM, Erlandsen SL. Presence of human giardia in domestic, farm and wild animals, and environmental samples suggests a zoonotic potential for giardiasis. Vet Parasitoly 2002; 108: 97-107.
- Asher A, Hose G, Power M. Giardiasis in nsw. Identification of giardia duodenalis assemblages contributing to human and cattle cases, and an epidemiological assessment of sporadic human giardiasis. Infect Genet Evol 2016; 44: 157-161.
- Sadeghi H, Borji H. A survey of intestinal parasites in a population in qazvin, north of iran. Asian Pac J Trop Di 2015; 5: 231-233.
- Nasiri V, Esmailnia K, Karim G, Nasir M, Akhavan O. Intestinal parasitic infections among inhabitants of karaj city, tehran province, iran in 2006-2008. Korean J Parasitol 2009; 47: 265-268.
- Salman YJ, Al-Taee A-RA, Abid AM. Prevalence of giardia lamblia among iraqi displaced peoples in kirkuk province. Int J Curr Microbiol App Sci 2016; 5: 753-760.
- Arani AS, Alaghehbandan R, Akhlaghi L, Shahi M, Lari AR. Prevalence of intestinal parasites in a population in south of Tehran, Iran. Rev Inst Med Trop Sao Paulo 2008; 50: 145-149.
- Taherkhani H, Shariati S, Abdolahi N, Roshandel G. Clinical manifestations of giardiasis in iran. J Clin Diagn Res 2009; 3: 1416-1418.
- Fallah E, Nahavandi KH, Jamali R, Poor BM, Asgharzadeh M. Molecular identification of giardia duodenalis isolates from human and animal reservoirs by pcr-rflp. J Biol Sci 2008; 8: 1-6.
- Nahavandi KH, Fallah E, Asgharzadeh M, Mirsamadi N, Mahdavipour B. Glutamate dehydrogenase and triose-phosphate-isomerase coding genes for detection and genetic characterization of giardia lamblia in human feces by pcr and pcr-rflp. Turk J Med Sci 2011; 41: 283-289.
- Roointan ES, Rafiei A, Samarbaf-Zadeh AR, Shayesteh AA, Shamsizadeh A, Borujeni MP. Genotype analysis of giardia lamblia isolated from children in ahvaz, southwest of iran. Jundishapur J Microbiol 2013; 6: 279-283.
- Al-Mohammed HI. Genotypes of giardia intestinalis clinical isolates of gastrointestinal symptomatic and asymptomatic saudi children. Parasitol Res 2011; 108: 1375-1381.
- Read C, Walters J, Robertson ID, Thompson RC. Correlation between genotype of Giardia duodenalis and diarrhoea. Int J Parasitol 2002; 32: 229-231.
- Foronda P, Bargues MD, Abreu-Acosta N, Periago MV, Valero MA. Identification of genotypes of Giardia intestinalis of human isolates in Egypt. Parasitol Res 2008; 103: 1177-1181.
- Traub R, Monis P, Robertson I, Irwin P, Mencke N, Thompson R. Epidemiological and molecular evidence supports the zoonotic transmission of giardia among humans and dogs living in the same community. Parasitology 2004; 128: 253-262.
- Mahdy AM, Surin J, Wan KL, Mohd-Adnan A, Al-Mekhlafi MH, Lim Y. Giardia intestinalis genotypes: Risk factors and correlation with clinical symptoms. Acta Tropica 2009; 112: 67-70.
- Amar C, Dear P, Pedraza-Diaz S, Looker N, Linnane E, McLauchlin J. Sensitive pcr-restriction fragment length polymorphism assay for detection and genotyping of giardia duodenalis in human feces. J Clin Microbiol 2002; 40: 446-452.
- Haque R, Roy S, Kabir M, Stroup SE, Mondal D. Giardia assemblage A infection and diarrhea in Bangladesh. J Infect Dis 2005; 192: 2171-2173.
- Sarkari B, Ashrafmansori A, Hatam GR, Motazedian MH, Asgari Q. Genotyping of Giardia lamblia isolates from human in southern Iran. Trop Biomed 2012; 29: 366-371.
- Pestehchian N, Rasekh H, Babaei Z, Yousefi HA, Eskandarian AA, Kazemi M, Akbari M. Identification of genotypes of giardia duodenalis human isolates in isfahan, iran, using polymerase chain reaction-restriction fragment length polymorphism. Adv Biomed Res 2012; 1: 84.
- Eckert J. New aspects of parasitic zoonoses. Vet Parasitol 1989; 32: 37-55.
- Organization WH: Parasitic zoonoses. Report of a who expert committee, with the participation of fao [meeting held in geneva from 14 to 20 november 1978]. 1979 .
- Palmer SR, Soulsby L, Simpson DIH. Zoonoses: Biology, clinical practice and public health control. Oxford university press 1998.
- Sprong H, Cacciò SM, van der Giessen JW; ZOOPNET network and partners. Identification of zoonotic genotypes of Giardia duodenalis. PLoS Negl Trop Dis 2009; 3: e558.
- Hoque ME, Hope VT, Kjellström T, Scragg R, Lay-Yee R. Risk of giardiasis in Aucklanders: a case-control study. Int J Infect Dis 2002; 6: 191-197.
- Warburton A, Jones P, Bruce J. Zoonotic transmission of giardiasis. A case control study. Communicable Dis Rep CDR Rev 1994; 4: R32-36.
- Alyousefi NA, Mahdy MA, Xiao L, Mahmud R, Lim YA. Molecular characterization of Giardia duodenalis in Yemen. Exp Parasitol 2013; 134: 141-147.
- Liu A, Zhang X, Zhang L, Wang R, Li X, Shu J, Zhang X, Shen Y, Zhang W, Ling H. Occurrence of bovine giardiasis and endemic genetic characterization of giardia duodenalis isolates in heilongjiang province, in the northeast of china. Parasitol Res 2012; 111: 655-661.
- Helmy MM, Abdel-Fattah HS, Rashed L. Real-time pcr/rflp assay to detect giardia intestinalis genotypes in human isolates with diarrhea in egypt. J Parasitol 2009; 95: 1000-1004.