ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 10
Cloning and sequence analysis of the full-length genome of Japanese encephalitis virus JEV-YIN strains
1Henan University of traditional Chinese Medicine, No.1 Jinshui Road Zhengzhou Henan, PR China
2Wuhan Zhijun IT limited company, Wuhan, PR China
3Liaoning University, No.66, Chongshan mid Road, Huanggu District, Shenyang, Liaoning, PR China
#These authors contributed equally to the work
- *Corresponding Authors:
- Chunyu Zhu
Liaoning University, PR China
- Fuchun Si
Henan University of traditional Chinese Medicine
PR China
Accepted on February 28, 2017
Japanese Encephalitis Virus (JEV) is a mosquito-borne flavi virus that causes severe acute viral encephalitis in humans. JEV exists in a zoonotic cycle between mosquitoes and pigs and/or water birds. JEV is a member of the genus Flaviviruses of the Flaviviridae family. The genomic RNA of JEV is a positive-sense, single-stranded RNA. In this study, the primer was studied by the method of segmented design according to the whole sequence (Genebank sequence number M18370) of the genebank JaOArS982 Japanese encephalitis virus strains. The material was gained from Hubei academy of agricultural sciences’ cultured cells. RNA was extracted from the cells and then subjected to RT-PCR, which yielded ten different segments. The complete nucleotide and deduced amino acid sequences of the JEV strain JEV-Yin were determined. The sequenced genome of JEV-Yin was 10,976 nucleotides in length. Sequence comparison of the JEV-Yin polyprotein Open Reading Frame (ORF) with those of 33 other JEV strains revealed that the nucleotide sequence divergence ranged from 0.53% to 25.14%. Sequence analysis of the full-length JEV-Yin E gene sequence with those of 34 other JEV isolates also identified nucleotide divergence, ranging from 0.33% to 22.27%. Phylogenetic analyses indicated that the JEV-Yin strain belonged to genotype III. These results may provide insight into the molecular properties and developing infectious cDNA clone of JEV-Yin strain.
Keywords
Japanese Encephalitis Virus (JEV), Full-length genome, Sequence analysis, JEV-Yin strain.
Introduction
Japanese Encephalitis Virus (JEV) is a mosquito-borne flavivirus that causes severe acute viral encephalitis in humans. JEV exists in a zoonotic cycle between mosquitoes and pigs and/or water birds [1]. In development of JE, pigs are considered as the most important natural amplifying host among several animals. JEV is a member of the genus Flaviviruses of the Flaviviridae family. The genomic RNA of JEV is a positive-sense, single-stranded RNA that is capped at the 5’-end and unpolyadenylated at the 3’-end. The genome contains a single long Open Reading Frame (ORF) that encodes a polyprotein flanked by 5’ and 3’ Non-Translated Regions (NTRs). The full-length genome consists of 10976 bp, which includes a 95 bp 5’-Untranslated Regions (UTRs), a 595 bp 3’-untranslated region. The three structural proteins, Capsid (C), membrane (M; a mature form of its precursor protein, prM), and Envelope (E), and the seven non-structural proteins, designated NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5, are encoded 3433 amino acids altogether [2]. Recently phylogenetic analyses have mainly focused on partial sequences derived from either the prM or E gene, which is important in several biological activities, including hemagglutination, neutralization, viral binding to cellular receptors and membrane fusion [3]. Partial sequence analysis of the prM gene indicates that JEV strains can be subdivided into four genotypes [4], then the E region was targeted for JEV phylogenetic analyses and JEV strains were divided into five genotypes [5-7].
In China, JE epidemics have occurred for over 50 years since the first JE case was reported in 1949. JEV is active in all of the provinces in China except for Tibet, Xinjiang, and Qinghai, and nearly 80% of globally reported cases occur in China [7]. A number of geographically diverse JEV strains have been isolated at different times from humans, mosquitoes and pigs. Lots of isolates of JEV strain were obtained from China and their genomes were partially sequenced, but only several strains have been fully sequenced.
In this study, the complete nucleotide and deduced amino acid sequences of the JEV-Yin strain, isolated from Hubei Agriculture academy of Science, China, were determined. By comparing the full-length genome of the JEV-Yin strain with all other fully sequenced JEV strains currently available, we have fully characterized it at the molecular level and established its relationship to the other 33 fully sequenced JEV strains. We also discuss the genetic relationship of the JEV-Yin strain to a large and heterogeneous selection of JEV strains, isolated from different geographic regions worldwide at different periods, and the results showed that JEV-Yin strain belongs to genotype III. These results may provide insight into the molecular properties and developing infectious cDNA clone of JEV-Yin strain.
Materials and Methods
Material
Brain tissue of pig infected with JEV was provided by the Hubei Agriculture academy of Science.
Escherichia coli DH5a was stored in our own laboratory.
Reagent
TRIZOL were purchase from SIGMA, various tools enzyme were purchase from TakaRa, M-MLV reverse transcriptase, TaqTM, Ribonuclease Inhibitor, DNA marker, pGEM-T-Easy Vector kit were purchase from Promega. KOD-Plus was purchase from TakaRa.
Primer design and synthesis
PCR primers were block designed according to Japanese encephalitis virus JaOArS982 strains of whole sequence (Genebank serial number M18370), synthesized by Beijing aoke Biotechnology Company (Table 1).
| Name of prime | Sequence of prime | Site length (bp) |
|---|---|---|
| JEV1F | 5’aga agt tta tct gtg tga act tct tgg c | 1-28 |
| JEV1R | 5’aag gag cat tgg gtg tta ttg taa ac | 1500-1479 1500 |
| JEV2F | 5’caa cga gaa gcg agc tga tag tag | 1220-1243 |
| JEV2R | 5’caa gac att ccc cca aag agt g | 2350-232 1130 |
| JEV3F | 5’gaa ggg agc tca aag act ggc agc | 2210-2233 |
| JEV3R | 5’ gtg gcc aga aac atc acc aga ag | 3592-3570 1382 |
| JEV4F | 5’caa tgg tga aat ggt tga ccc | 3536-3556 |
| JEV4R | 5’ccg ttc aag cca cat atc tgt tg | 4382-4360 846 |
| JEV5F | 5’gtc aat acc ctt cat gct ggc a | 4301-4322 |
| JEV5R | 5’gca ttt gag tca gga aaa gga tcc | 5599-5576 1298 |
| JEV6F | 5’acc acg gat cct ttt cct gac tca | 5571-5594 |
| JEV6R | 5’ctt tcc agt tca ggc aca tca gtg | 7402-7379 1831 |
| JEV7F | 5’agc ggc tgg aat aat gaa gaa tg | 7334-7356 |
| JEV7R | 5’cag cgg ctc cac taa ccc aat aca | 8355-8332 1021 |
| JEV8F | 5’ggc tag tgc gtc tcc ccc tgt c | 8290-8300 |
| JEV8R | 5’cac tcc ccc ttt gat ctt ctc ttg | 9489-9466 1199 |
| JEV9F | 5’tga cac cgc cgg atg gga cac tag | 9281-9304 950 |
| JEV10 (F) | 5’cgt acg tgg gaa agc gtg agg aca | 10201-10224 |
| JEV9, 10R | 5’gta ggt acc aga tcc tgt gtt ctt cct cac cac cag | 10976-10950 775 |
Table 1. The primers used in the experiment, the position and the length of the fragments amplified.
Preparation of virus and extracted viral genomic RNA
The JEV-Yin strains were propagated in DH5α cell and the infected culture fluid was frozen and thawed three times. After centrifugation, the supernatant was stored at -70°C until use. Total RNA was extracted from Japanese encephalitis virus of cell cultures with TRIZOL method. To isolate RNA, Japanese encephalitis virus of cell cultures (200 μl) was added into trizol, homogenated and rested at room temperature for 5 minutes.
RT-PCR and Whole-genome sequencing
We mixed the purified RNA (3 μg) with 1 μL Random primers and added Diethylpyrocarbonate (DEPC) water to obtain a final volume of 15 L. Then, the mixture was heated at 70°C for 5 min and immediately chilled by placing on ice. Subsequently, we added 3 μL of 5X Moloney Murine Leukemia Virus (MMLV) buffer (Promega, USA), 5 μL dNTP (10 mM; TaKaRa, Japan), 1 μL RNase inhibitor (Promega, USA), 1 μL M-MLV Reverse Transcriptase (200 μ/μl; Promega, USA), and DEPC water to bring the final volume of the solution to 25 μL. The reaction mixture was incubated at 37°C for 60 min, received cDNA. Polymerase Chain Reaction (PCR) was performed using cDNA as the template, using designed primer (Table 1). PCR was performed using 5 μl cDNA, 1 μl each primer, 1 μl KOD-Plus, 2 μl MgSO4, 5 μl 10X KOD buffer and 2 mM dNTP 4 μL in a total volume of 50 μL. The PCR cycles were 94°C for 5 min; 35 cycles at 94°C for 45 sec, 42°C for 45 sec, and 72°C for n min (extended time vary from the different fragment length); and a final extension at 72°C for 10 min; the products were stored at 4°C.
The amplified fragments of PCR were purified using a gel extraction kit (Tiangen, Beijing, China); then, the purified fragments were cloned into pGEM-T-Easy Vector (TaKaRa, Japan). The connection system was performed using 5 μL 2X Rapid Ligation Buffer, 1 μL T-Easy vector, 1 μL T4DNA Ligase, 3 μL PCR product. Then keep it one hour at room temperature and connect it overnight at 4°C. The connection product were electrotransformed in Escherichia coli DH5α Then picked the white colonie, The plasmid DNA that white colonie was extracted by Small amounts of plasmid extraction kit. The results were identified by EcoRI or NotI Restriction and double digestion products obtained were confirmed by sequencing, which were sent to Shanghai yabo Biotechnology Company.
Multiple alignments and phylogenetic analysis
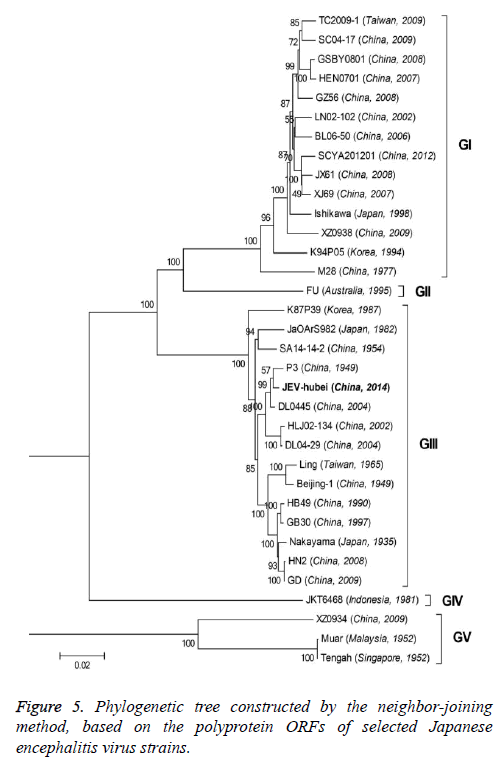
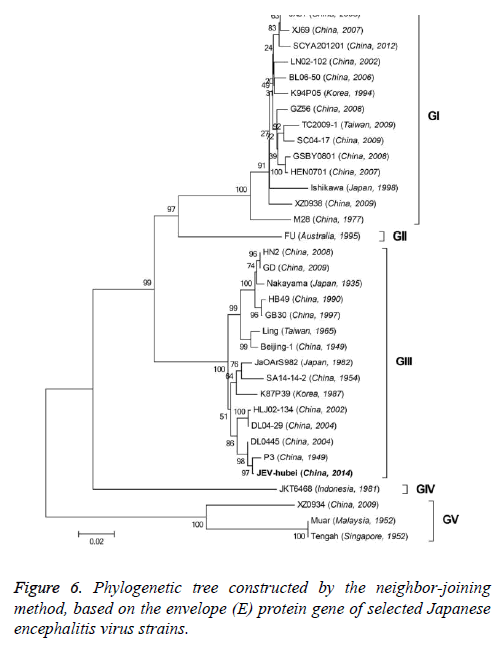
The 33 JEV strains used in multiple alignments and phylogenetic analysis for which complete sequences are presently available from the NCBI nucleotide sequence databases (including the JEV-Yin strain). In the case of the viral E gene, our analysis was performed with a total of 34 strains that were available in the GenBank. Multiple sequence alignments and sequence similarity calculations between aligned nucleotide and amino acid sequences were performed using computer software program (DNASTAR Inc., Madison, WI, USA). The phylogenetic trees were reconstructed on aligned nucleotide sequences by using the ClustalV method. The genomic sequence of 33 strains and the E gene sequence of 34 strains were employed in the generation of the phylogenetic trees. For construction of rooted trees, the Murray Valley Encephalitis virus (MVE) genome was used as out group in all analyses. Bootstrap resampling analysis of 1000 replicates was used to assess confidence values of virus groupings. All trees were drawn using the Neighbour-Joining (NJ) method in MEGA6 software.
Results
RT-PCR amplification results
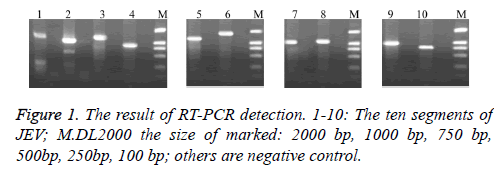
We can receive the cDNA by reverse transcription using random primers after RNA was extracted from JEV. Then PCR was amplified using our own design synthesis of segmented primers. Electrophoresis results showed that the resulting fragments is consistent with the expected results (Figure 1).
Cloning of PCR products
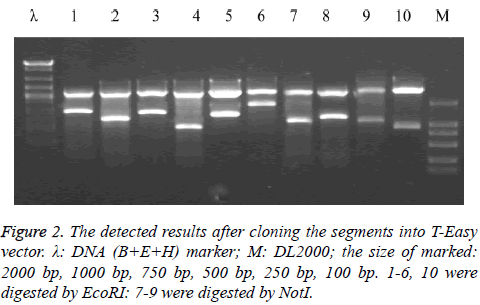
The above 10 pieces of PCR amplification products were cloned to T-Easy vector and were identified using EcoRI or NotI enzyme and preliminary judgment is correct from the enzyme identification of electrophoresis (Figure 2).
Full-length nucleotide and deduced amino acid sequence analysis
The full-length RNA genome of JEV-Yin is 10976-nucleotides in length which includes a 95 bp 5’-Untranslated Region (UTR), a 595 bp 3’-untranslated region. In order to characterize the molecular structure of the JEV-Yin genome and to determine how it is related to other fully sequenced JEV strains, we compared the complete JEV-Yin genomic sequence with sequences of 33 JEV strains available in GenBank (Table 2).
| Strain name | Country | Year | Source | Genotype | GenBank accession no. |
|---|---|---|---|---|---|
| GZ56 | China | 2008 | Human | I | HM366552 |
| M28 | China | 1977 | Mosquito | I | JF706279 |
| K94P05 | Korea | 1994 | Mosquito | I | AF045551 |
| Ishikawa | Japan | 1998 | Mosquito | I | AB051292 |
| LN02-102 | China | 2002 | Mosquito | I | JF706278 |
| BL06-50 | China | 2006 | Mosquito | I | JF706270 |
| XJ69 | China | 2007 | Mosquito | I | EU880214 |
| GSBY0801 | China | 2008 | Mosquito | I | JF706274 |
| SC04-17 | China | 2009 | Mosquito | I | GU187972 |
| XZ0938 | China | 2009 | Mosquito | I | HQ652538 |
| TC2009-1 | Taiwan | 2009 | Mosquito | I | JF499790 |
| HEN0701 | China | 2007 | Swine | I | FJ495189 |
| JX61 | China | 2008 | Swine | I | GU556217 |
| SCYA201201 | China | 2012 | Swine | I | KM658163 |
| FU | Australia | 1995 | Human | II | AF217620 |
| HB49 | China | 1990 | Bat | III | JF706284 |
| GB30 | China | 1997 | Bat | III | FJ185037 |
| HN2 | China | 2008 | Bat | III | JN711459 |
| GD | China | 2009 | Bat | III | JN711458 |
| Nakayama | Japan | 1935 | Human | III | EF571853 |
| Beijing-1 | China | 1949 | Human | III | L48961 |
| P3 | China | 1949 | Human | III | U47032 |
| Ling | Taiwan | 1965 | Human | III | L78128 |
| JaOArS982 | Japan | 1982 | Mosquito | III | M18370 |
| K87P39 | Korea | 1987 | Mosquito | III | AY585242 |
| HLJ02-134 | China | 2002 | Mosquito | III | JF706276 |
| DL04-29 | China | 2004 | Mosquito | III | JF706272 |
| DL0445 | China | 2004 | Mosquito | III | JN381854 |
| SA14-14-2 | China | 1954 | Vaccin strain | III | AF315119 |
| JKT6468 | Indonesia | 1981 | Mosquito | IV | AY184212 |
| Muar | Malaysia | 1952 | Human | V | HM596272 |
| Tengah | Singapore | 1952 | Human | V | KM677246 |
| XZ0934 | China | 2009 | Mosquito | V | JF915894 |
Table 2. Background of Japanese encephalitis virus strains used in this study.
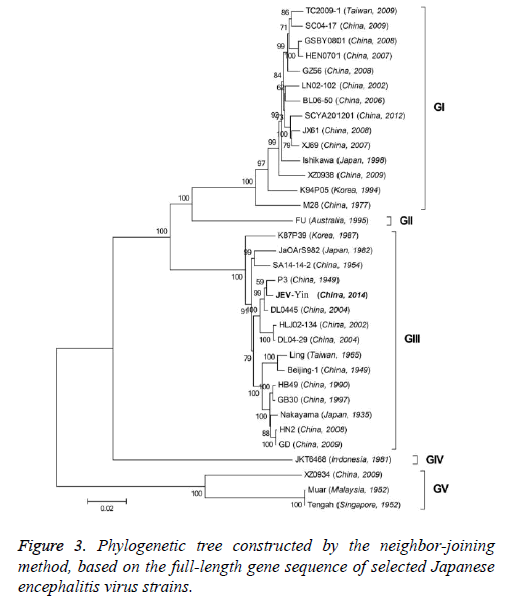
Phylogenetic analyses
To better understand the genetic relationships and evolution of JEV strains, we performed a phylogenetic analysis of the 33 fully sequenced JEV strains, including the JEV-Yin strain (Figure 3). This analysis showed that JEV-Yin strain was still closely related to that of P3 strain. Phylogenetic comparisons based on single JEV genes, including the 5’-NTR and 3’-NTR, showed that the E/prM gene-based phylogenetic tree corresponds well to the full-length genome-based tree, and both exhibit two major distinct clusters with several minor branches. When comparing its ORF with that of 33 other JEV strains isolated from different geographic regions or periods, high levels of similarity were observed, with nucleotide divergence ranging from 0.53% to 25.14%, and amino acid divergence ranging from 0.44% to 9.28%. A comparison of the JEV-Yin polyprotein ORF with that of SA14-14-2, the attenuated JEV vaccine strain currently widely used in China, discovered a total of 211 nucleotide differences (2.05%) and 50 amino acid differences (1.46%).
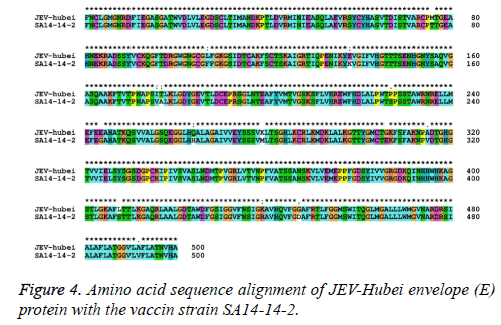
The nucleotide and amino acid sequences of the JEV-Yin strain E region were compared with those of other isolates available in GenBank. This analysis revealed that nucleotide divergence ranged from 0.33% to 22.27%, while deduced amino acid divergence ranged from 0.40% to 9.00%. A comparison of the JEV-Yin strain E gene with that of SA14-14-2 also indicated 36 nucleotide changes (2.40%), resulting in 15 amino acid changes (3.00%): E76 (Thr to Met), E107 (Phe to Leu), E138 (Lys to Glu), E176 (Val to Ile), E177 (Ala to Thr), E227 (Ser to Pro), E244 (Gly to Glu), E264 (His to Gln), E279 (Met to Lys), E306 (Glu to Gly), E315 (Val to Ala), E408 (Ser to Lys), E439 (Arg to Lys) and E492 (Val to Ala). Amino acid sequence alignment of JEV-hubei Envelope (E) protein with the vaccine strain SA14-14-2 (Figure 4).
On the basis of the ORF gene sequences of 33 JEV isolates and the E gene sequences of 34 JEV isolates, phylogenetic trees were constructed (Figures 5 and 6). Both phylogenetic trees provided similar topology, and the analysis indicated that the JEV-Yin strain belonged to genotype III. On the basis of 240 nucleotide of the C/prM gene, JEV strains were divided into five genotypes two decades ago [8,9]. However, the functional significance of the prM genetic variations remains to be established, and genetic relationships based on short sequences (<300 nt) should be required to take into account the biological significance and may be considered with caution [10,11]. Subsequently, the full-length E gene of JEV and its deduced protein sequence have been used as reliable phylogenetic markers, given that the E protein plays an important role in the JEV life cycle [1]. In China, many JEV strains belonging to genotype I and genotype III have been reported, and it was suggested by surveillance data that genotype I strains have been gradually replacing genotype III as the dominant strains in Asia. In this study, phylogenetic analyses of the JEV-Yin strain indicated it belonged to genotype III.
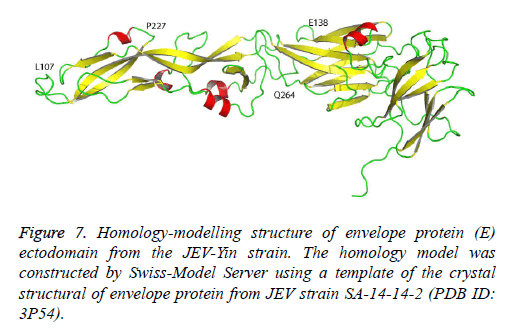
The crystallographic structure of the E protein of the flavivirus tick-borne encephalitis virus revealed that the E protein consists of three domains. The difference in the E gene of JEV strains, particularly in domain III [12], may contribute to the diversity of neutralizing and antigenic properties [13]. Previous research has also indicated that mutations in domain III modulated virus binding and entry into host cells. The JEV-Yin strain homology model was constructed by Swiss-Model Server using a template of the crystal structural of envelope protein from JEV strain SA-14-14-2 (PDB ID: 3P54) (Figure 7). In this study, comparison of the E gene of JEV-Yin strain with the vaccine strain SA14-14-2 revealed 15 amino acid substitutions. Among these variations, the Glu to Lys alteration at position 138 is consistent with previous observations that the E138 mutation is associated with attenuation of the JEV strain and inhibits viral spread from cell to cell [14]. Other groups have also demonstrated that mutations of residues E138, E306 and E389 of the E protein reduce the efficiency of viral binding to heparan sulphate residues on target cells [5]. In our results, a substitution (Met to Lys) was observed at the position E279, which may increase virulence in mice [15].
In this study, phylogenetic analyses of the JEV-Yin ORF and the ORF of 33 other JEV strains isolated from several countries revealed the lowest nucleotide divergence and the lowest amino acid divergence with the P3 strain. Meanwhile, phylogenetic analyses of its E gene and the E gene of 34 other JEV strains revealed the lowest nucleotide divergence and the lowest amino acid divergence with the P3 strain.
Discussion
We have determined the complete nucleotide and deduced amino acid sequences of the JEV-Yin strain, isolated from Hubei Agriculture academy of Science, China. The genome sequence of JEV-Yin was 10,976 nucleotides in length. Phylogenetic analyses indicated that the JEV-Yin strain belonged to genotype III.
In previous studies, a number of available JEV strains have been partially sequenced for prM and E gene regions, which are important for induction of protective immunity [10]. The viral E protein has been demonstrated to be a reliable phylogenetic marker. The E protein plays an important role in tissue tropism, cell fusion and infection, virus maturation, and protection [15,16]. Its corresponding protein established phylogenetic markers for JEV [6,17,18] New JEV isolate JEVYin genomic RNAs showed a nucleotide sequence similar to genotype III. But the live vaccine strain SA14-2-2 in China also belongs to genotype III.
In China, the first JEV genotype I strain was isolated from Yunnan Province in 1979, then the genotype I strains were separately isolated from Shanghai City, Liaoning and Henan provinces from 1982 to 2005. But in the same period, the JEV genotype III strains were isolated from Heilongjiang, Yunnan, Guizhou, and Fujian provinces [7,19]. It showed that the JEV genotype I and III strains were coexistent in the vast territory of China at present, but the distribution region within China of two genotypes JEV had also changed. The reasons for emergence of genotype I in China are uncertain, but may include changes of in agricultural practice, animal husbandry and distribution of migrating birds such as heron and egret [7,19,20] so grasping the changes in the distribution of the different genotype of the JEV in due time will play an important role in prevention and treatment of JE.
Compared with JEV-Yin and SA14-14-2 strains, the homology of nucleotide sequence in E gene was 87.8%; but the homology of amino acid was 97.0%. This showed that nucleotide difference of the envelope gene regions commonly located at the third site of amino acid codon, and silence mutate was the most mode in mutate sites. The previous researches [12,21] showed that the three dimensional structure of the JEV envelope glycoprotein composed chiefly of three domains, Domain I: E1-E51, E137-E196, E293-E311; Domain II: E52- E137, E197-E292; Domain III: E310-E411. These domains determine the virus’s hemagglutination ability, cellular tropism, viral virulence, and induction of protective immune response. So contrasting with the vaccine strain SA-14-14-2, we analysed the amino acid residue in the active region (E1-E411) of E protein of the isolate JEV-Yin. The results showed that there were amino acid substitutions in 10 sites of the active domains, including E76, E138, E176, E177 of Domain I; E107, E227, E244, E264, E279 of Domain II; and E306 of Domain III. The previous research demonstrated that, disulfide linkage was the necessary structure for antigen-antibody fixation, which was synthesized by two cysteine located on E304 and E335 of the E protein [8]. The research results about continuous polypeptide segments (E307-E309, E327-E333, E386-E390) in Domain III of JEV E protein showed that three residues forming the novel cis-proline turn structure were all important in eliciting JEV-specific neutralizing antibodies. But the polypeptide segments in all these sites of JEV-Yin were not mutated. So this results and the higher homology of amino acid suggested that the protection of vaccine strain SA14-14-2 was able to theoretically cover JEV including the isolate from Shaanxi Province.
Characterization of the JEV-Yin strain at molecular level would be an important step towards identifying those properties of the virus, which may aid in the design and construction of recombinant JEV vaccine that are based on the E protein. Future studies may be aimed to investigate the efficacy of current JEV vaccine against the JEV-Yin strain.
Acknowledgements
This study is supported by the Henan Provincial Department of Science and Technology Research Project (No. 162102310465), 2015 Key scientific research project of Henan higher education (No. 15A310020) and Nursery Engineering of Henan traditional Chinese Medicine University (No. MP2015-05). We also thank the Hubei Agriculture academy of Science for providing the materials.
Disclosure of Potential Conflicts of Interest
The authors declare that they have no conflict of interest.
Research Involving Human Participants and/or Animals
For this type of study formal consent is not required.
References
- Monath TP, Heinz FX. Flaviviruses. Fields Virology Lippincott-Raven: Philadelphia (3rd edn) 1996; 961-1034.
- Wang WH, Zhang YM, Xin-Gang XU, Xing FS, Tang QH. Cloning And Sequence Analysis Of The Full-Length Genome Of Japanese Encephalitis Virus Strain Sxbj07 Isolated From Swine. Agr Sci China 2009; 8: 1392-1402.
- Gritsun TS, Venugopal K, Zanotto PM, Mikhailov MV, Sall AA. Complete sequence of two tick-borne flaviviruses isolated from Siberia and the UK: analysis and significance of the 5 and 3-UTRs. Virus Res 1997; 49: 27-39.
- Chen WR, Tesh RB, Rico-Hesse R. Genetic variation of Japanese encephalitis virus in nature. J Gen Virol 1990; 71: 2915-2922.
- Mangada MNM, Takegami T. Molecular characterization of the Japanese encephalitis virus representative immunotype strain JaGAr 01. Virus Res 1999; 59: 101-112.
- Solomon T, Ni H, Beasley DW, Ekkelenkamp M, Cardosa MJ. Origin and evolution of Japanese encephalitis virus in southeast Asia. J Virol 2003; 77: 3091-3098.
- Wang HY, Takasaki T, Fu SH, Sun XH, Zhang HL, Wang ZX. Molecular epidemiological analysis of Japanese encephalitis virus in China. J Gen Virol 2007; 88: 885-894.
- Ali A, Igarashi A. Antigenic and genetic variations among Japanese encephalitis virus strains belonging to genotype 1. Microbiol Immunol 1997; 41: 241-252.
- Huong VT, Ha DQ, Deubel V. Genetic study of Japanese encephalitis viruses from Vietnam. Am J Trop Med Hyg 1993; 49: 538-544.
- Ni H, Barrett AD. Nucleotide and deduced amino acid sequence of the structural protein genes of Japanese encephalitis viruses from different geographical locations. J Gen Virol 1995; 76: 401-407.
- Bennett SN, Gubler DJ, Ooi EE, Vasudevan S, Farrar J. Taxonomy and evolutionary relationships of flaviviruses CAB eBooks 2014; 322-333.
- Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 1995; 375: 291-298.
- Gritsun TS, Holmes EC, Gould EA. Analysis of flavivirus envelope proteins reveals variable domains that reflect their antigenicity and may determine their pathogenesis. Virus Res 1995; 35: 307-321.
- Zhao Z, Date T, Li Y, Kato T, Miyamoto M, Yasui K. Characterization of the E-138 (Glu/Lys) mutation in Japanese encephalitis virus by using a stable, full-length, infectious cDNA clone. J Gen Virol 2005; 86: 2209-2220.
- Monath TP, Arroyo J, Levenbook I, Zhang ZX, Catalan J, Draper K. Single mutation in the flavivirus envelope protein hinge region increases neurovirulence for mice and monkeys but decreases viscerotropism for monkeys: relevance to development and safety testing of live, attenuated vaccines. J Virol 2002; 76: 1932-1943.
- Kimura-Kuroda J, Ichikawa M, Ogata A, Nagashima K, Yasui K. Specific tropism of Japanese encephalitis virus for developing neurons in primary rat brain culture. Arch Virol 1993; 130: 477-484.
- Williams DT, Wang LF, Daniels PW, Mackenzie JS. Molecular characterization of the first Australian isolate of Japanese encephalitis virus, the FU strain. J Gen Virol 2000; 81: 2471-2480.
- Nga PT, del Carmen Parquet M, Cuong VD, Ma SP, Hasebe F, Inoue S. Shift in Japanese encephalitis virus (JEV) genotype circulating in northern Vietnam: implications for frequent introductions of JEV from Southeast Asia to East Asia. J Gen Virol 2004; 85: 1625-1631.
- Chen D, Wang HY, Fu SH, Song H, Deng J, Yang YL. Molecular characteristics of three new isolates of Japanese encephalitis virus in Fujian Province (in Chinese). Chin J Exp Clin Virol 2005; 19: 5-8.
- Li XY, Song H, Fu SH, Wang HY, Yu YX, Dong GM. The molecular biology of Japanese encephalitis viruses isolated in China 20. Chin J Gen Virol 2004; 20: 200-209.
- Kolaskar AS, Kulkarni-Kale U. Prediction of three-dimensional structure and mapping of conformational epitopes of envelope glycoprotein of Japanese encephalitis virus. Virology 1999; 261: 31-42.