ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 17
Clinical significance of miRNA-433 expression in hepatocellular carcinoma
Zhanzhan Li1, Yanyan Li2* and Jing Xue3
1Department of Oncology, Xiangya Hospital, Central South University, Changsha, Hunan Province, PR China
2Department of Outpatient, Xiangya Hospital, Central South University, Changsha, Hunan Province, PR China
3Departmeng of Archive, Xiangya Hospital, Central South University, Changsha, Hunan Province, PR China
- *Corresponding Author:
- Yanyan Li
Department of Outpatient
Xiangya Hospital
Central South University, PR China
Accepted on August 18, 2017
Background: Previous studies found miRNAs can play important role in the pathogenesis, metastasis and prognosis of hepatocellular carcinoma by regulating the expression of its target genes. We evaluated the clinical significance of miRNA-433 expression in hepatocellular carcinoma.
Method: We collected 28 primary hepatocellular carcinoma specimens matched by 28 normal paracarcinoma tissues. The RNA concentration was calculated by the following formula: OD260 × 50 times × 40/100. We detected the absorbance at 260 nm and 280 nm, and calculated OD260/OD280. The purity was considered to be acceptable when the ratio value fallen into between 1.8 and 2.0. The expression of MiR-433 was analysed via Real time-PCR method. The Cq value was calculated, and Data were processed using 2-ΔΔCT method.
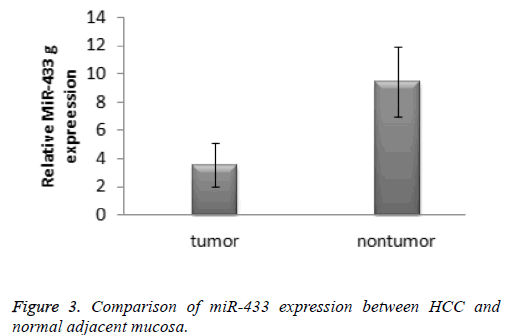
Results: According to the 2-ΔΔCT method, the expression of miR-433 in the hepatocellular carcinoma specimens was significantly lower than that in the normal tissue specimens (3.61 ± 1.35 vs. 9.39 ± 2.46, t=-10.899, P<0.001). The receiver operating characteristic cure was used to identify the diagnostic value of miR-433 expression levels in hepatocellular carcinoma. The area under curve of miR-433 was 0.770 (95% confidence interval: 0.594-0.947).
Conclusion: The low expression of miR-433 in hepatocellular carcinoma may be related to occurrence of hepatocellular carcinoma. miR-433 expression might have a moderately diagnostic value to hepatocellular carcinoma.
Keywords
miRNA, Hepatocellular carcinoma, miR-433.
Introduction
Primary Hepatocellular Carcinoma (HCC) is one of the most common malignant tumors around the world. According to the report of the China’s official cancer registry, the mortality of HCC ranked third in all cancer mortality in China, and 90% of death was due to metastasis [1,2]. Previous studies have shown that the proliferation and invasion mechanisms of HCC involved genetic mutations including oncogene activation, tumor suppressor gene inactivation, gene mutation and overexpression of genes as well as several signal pathways [3]. However, the specific molecular mechanism is not fully understood. Therefore, it is of great significance to explore the molecular mechanism of hepatocellular carcinoma, and to explore new intervention strategies for the treatment and prognosis of hepatocellular carcinoma. The expression of small miRNA can truly reflect the occurrence, development and prognosis of disease. The specific miRNA expression profile analysis of different types of tumors showed that there were specific markers of miRNA between HCC tissues and normal tissues as well as different types of liver cancer tissues [4]. MiRNA can play important role in the pathogenesis, metastasis and prognosis of HCC by regulating the expression of its target genes [5,6]. Tan found miR-433-3p had higher sensitivity and specificity in diagnosing HCC via area under the receiver operating characteristic curve evaluation [7]. Therefore, it is of great significance to explore the miRNA expression pattern in normal liver tissues and HCC cells, suggesting that the potential targets for the treatment of viral infectious diseases and HCC.
Materials and Methods
Samples collections
We collected 28 primary hepatocellular carcinoma specimens matched by 28 normal para-carcinoma tissues. All specimens were from those patients who underwent surgical resections in Xiangya Hospital of Central University. All specimens were stored in liquid nitrogen at -80ºC temperature for back up. This study was approved by the institutional review board of Xiangya Hospital.
RNA extraction and quantitative analyses
The extraction of RNA was according to the instruction of RNeasy FFPE Kit (NO.73504) from Qiagen Company (Qiagen, German). The extracted RNA was diluted by 50 times, and examined absorbance at 260 nm. The RNA concentration was calculated by the following formula: OD260 × 50 times × 40/100. We detected the absorbance at 260 nm and 280 nm, and calculated OD260/OD280. The purity was considered to be acceptable when the ratio value fallen into between 1.8 and 2.0. The total RNA was reversely transcribed into cDNA according to TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA, USA). The expression of miRNA-433 was analysed via real time-PCR method. The positive, negative and blank control experienced centrifugation 1000 r/min for 3 min, and were placed in Light Cycler 480 reaction plated according to the following procedure: 95°C for 5 min (pre-denaturation), 95°C for 10 s, 60°C for 10 s, 72°C for 10 s (40 cycles for PCR); 95°C for 5 s, 65°C for 60 s, 97°C for 0 s (melting curve analyses); 40°C for 30 s (cooling). The Cq value was calculated, and data were processed using 2-ΔΔCT method.
Statistical analysis
All statistical analyses were performed on Statistical Package for the Social Sciences version 19.0. Data was expressed by using mean ± standard deviation. The student t test was used to compare the expression of RNA between hepatocellular carcinoma specimens and normal para-carcinoma tissue. P<0.05 was considered as significant.
Results
Twenty-eight pairs of was completely extracted. The percent of purity pass was more than 99%. The total RNA was examined via spectrophotometer, the value of A260/A280 was from 1.8 to 2.0, which means the extracted RNA was complete and nondegradation. The extraction was suitable for detecting the expression of miRNA.
We used real-time quantitative PCR method to detect the expression of miR-433 through U6 gene control. As presented in Figures 1 and 2, the control and miR-433 melting cure were unimodal, and this showed that the specificity of primers were good. There was no interference of unprimed dimer. The logarithmic amplification stage, logarithmic amplification period and platform stage can be shown in the expansion curve of miRNA-433, which shows that the PCR products were abundant and sensitive to the primers.
According to the 2-ΔΔCT method, the expression of miR-433 in the hepatocellular carcinoma specimens was significantly lower than that in the normal tissue specimens (3.61 ± 1.35 vs. 9.39 ± 2.46, t=-10.899, P<0.001). The result was presented in the Figure 3. The receiver operating characteristic cure was used to identify the diagnostic value of miRNA-433 expression levels in hepatocellular carcinoma. The area under curve of miR-433 was 0.770 (95% confidence interval: 0.594-0.947).
Discussion
The occurrence of tumor was influenced by multifactorial, multi-gene, multi-step accumulation process. The most obvious difference from normal tissue is that the growth of tumor cells is self-directed, which means cell proliferation is out of control. Surgery is the primary method for hepatocellular carcinoma currently. Some issues existed for this method and others such as effectiveness and adverse prognosis. Therefore, it is quite urgent for clinical practice to find a new molecular biomarker for early diagnostic and treatment. In recent years, the abnormal expression of miRNA has been found in the process of digestive and cardiovascular diseases. The study of the regulation mechanism of miRNA on the tumor provides a new way for the diagnosis and treatment of diseases, and the relationship between the polymorphism of target gene and the disease is also worth of further discussion.
Previous studies reported the abnormal expression of some miRNAs was found in other types of tumor. Li reported there was significant difference between normal para-carcinoma tissue and gastric cancer tissue [8]. Luo found the expression of miRNA-433 was related to tumor stage, and clinical study showed some differences between early stage and end-stage tumor tissue [9]. Ueda also found the high expression level of miRNA-433 in patients with gastric cancer was related to overall survival, metastasis, tumor infiltration, and prognosis [10]. Luo reported that predicted 26 proteins targeted by miRNA-433, and found miRNA-433 could involve in the process of proliferation and invasion [11]. Our study found expression of miRNA-433 in the hepatocellular carcinoma specimens was significantly lower than that in the normal tissue specimens. Therefore, we hypothesized that miRNA-433 in tumor tissues may also control expression of some proteins in the form of negative regulation. Further research is needed.
The promotion of tumor cell apoptosis has become a hotspot in the research of anti-tumor therapy. But even if the cell apoptosis is blocked by drugs, cancer cells can still rely on autophagy to fight off metabolic stress and survive. Therefore, an ideal anti-tumor drug should both promote apoptosis and inhibit autophagy. Within the tumor cells of certain human impact the miRNA levels rise or fall, can make the tumor cell growth inhibition, aggressively weakened or even increased apoptosis, prompt gene expression levels of regulation mechanism of miRNAs research will be the new direction of targeted molecular treatment of clinical oncology. Previous study also found that miRNA-433 in liver cancer cell HepG2, SMMC7721 and BEL7404-7 below the expression in hepatocellular LO2 expression level. At the same time, through quantitative analysis in 28 cases of liver cancer tissues, we found that both expressed in liver cancer tissue present negative correlation, regulate the expression of miR-433 may be negative. Other studies have also confirmed that miRNA-433 can influence its transcriptional activity by targeting genes and regulate proliferation and migration of HCC cells [12,13]. This is the first reported role of miRNA-433 in HCC. Therefore, it can be speculated that miR-433 in HCC tissues and cells can also control the expression of some gene in the form of negative regulation. These results provide strong evidences for miRNAs in the study of tumor and other mechanisms of liver disease [14-16]. Our study has several limitations. First, our study was based on small sample size, and bias may exist. Second, our study was an exploratory research, we did not illustrate the mechanism, and we just give reasonable deduction. Further research was required. Finally, single miRNA was limited for complex diseases like cancer. It was reported that combined application of miRNA can improve the diagnostic ability. We may put these miRNA together for HCC early diagnostic. MiRNA should be combined with other traditional index or miRNAs for improving diagnostic ability. It was reported that miRNA-122 with AFP can increase the diagnostic ability of HCC. This point may give us some clues. Some study reported that miR-433 could inhibit proliferation of hepatocellular carcinoma cells by downregulation of PAK4 and upregulation of PI3K, AKT. It may be the possible mechanism [17,18].
In conclusion, the low expression of miR-433 in hepatocellular carcinoma may be related to occurrence of hepatocellular carcinoma. MiR-433 expression might have a moderately diagnostic value to hepatocellular carcinoma. This study was just preliminary discussion, and further research was needed.
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017; 67: 7-30.
- Malvezzi M, Carioli G, Rodriguez T, Negri E. Global trends and predictions in ovarian cancer mortality. Ann Oncol 2016; 27: 2017-2025.
- Tsai KW, Wu CW, Hu LY, Li SC, Liao YL. Epigenetic regulation of miR-34b and miR-129 expression in gastric cancer. Int J Cancer 2011; 129: 2600-2610.
- Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X, Zhou X, Gan J. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One 2014; 9: 107986.
- Wei H, Ke HL, Lin J, Shete S, Wood CG. MicroRNA target site polymorphisms in the VHL-HIF1α pathway predict renal cell carcinoma risk. Mol Carcinog 2014; 53: 1-7.
- Stahlhut C, Slack FJ. Combinatorial action of microRNAs let-7 and miR-34 effectively synergizes with erlotinib to suppress non-small cell lung cancer cell proliferation. Cell Cycle 2015; 14: 2171-2180.
- Tan Y, Ge G, Pan T, Wen D, Chen L, Yu X, Zhou X, Gan J. A serum microRNA panel as potential biomarkers for hepatocellular carcinoma related with hepatitis B virus. PLoS One 2014; 9: 107986.
- Li RD, Han GX, Li W, Tao KX. Down-regulation of microRNA-433 expression in gastric cancer: Possible mechanisms involved. World Chin J Dig 2012; 19: 1726-1731.
- Luo H, Zhang H, Zhang Z, Zhang X, Ning B. Down-regulated miR-9 and miR-433 in human gastric carcinoma. J Exp Clin Cancer Res 2009; 28: 82.
- Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol 2010; 11: 136-146.
- Luo HC. miRNAs expression profiliing ofgastric carcinoma and function of significantly down-regulated miR-9 and miR-433. Chongqing Med Univ 2010.
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin 2011; 61: 212-236.
- Tian Y, Yang W, Song J, Wu Y, Ni B. Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol Cell Biol 2013; 33: 2810-2816.
- Pan XP, Wang HX, Tong DM, Li Y, Huang LH, Wang C. miRNA-370 acts as a tumor suppressor via the downregulation of PIM1 in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci 2017; 21: 1254-1263.
- Shaker O, Alhelf M, Morcos G, Elsharkawy A. miRNA-101-1 and miRNA-221 expressions and their polymorphisms as biomarkers for early diagnosis of hepatocellular carcinoma. Infect Genet Evol 2017; 51: 173-181.
- Xie F, Yuan Y, Xie L, Ran P, Xiang X, Huang Q, Qi G, Guo X, Xiao C, Zheng S. miRNA-320a inhibits tumor proliferation and invasion by targeting c-Myc in human hepatocellular carcinoma. Onco Targets Ther 2017; 10: 885-894.
- Li ZZ, Shen LF, Li YY, Chen P, Chen LZ. Clinical utility of microRNA-378 as early diagnostic biomarker of human cancers: a meta-analysis of diagnostic test. Oncotarget 2016; 7: 58569-58578.
- Jun F, Hong J, Liu Q, Guo Y, Liao Y, Huang J, Wen S, Shen L. Epithelial membrane protein 3 regulates TGF-beta signaling activation in CD44-high glioblastoma. Oncotarget 2017; 8: 14343-14358.