ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2013) Volume 24, Issue 4
Asymmetric real-time PCR improves the SNP detection of VEGF+ 936C>T by melting curve analysis in LightCycler.
1Department of Medical Sciences, Conbuk National University Graduate School, 20, Geonji-ro, Jeonju, Chonbuk, 561- 712, Republic of Korea
2 Department of Biochemistry, Maharajgunj Medical Campus, Institute of Medicine, Tribhuvan University, Kathmandu, Nepal
3Department of Neurology, Chonbuk National University Medical School, 20, Geonji-ro, Jeonju, Chonbuk, 561-712, Republic of Korea
4Research Institute of Clinical Medicine of Chonbuk National University-Biomedical Research Institute of Chonbuk National University Hospital, 20, Geonji-ro, Jeonju, Chonbuk, 561-712, Republic of Korea
- *Corresponding Author:
- Byoung-Soo Shin
Department of Neurology, Chonbuk National
University Medical School, 20, Geonji-ro, Jeonju
Chonbuk, 561-712, Republic of Korea
Accepted August 22 2013
Citation: Yadav BK, Oh S-Y, Shin B-S. Asymmetric real-time PCR improves the SNP detection of VEGF+ 936C>T by melting curve analysis in LightCycler. Biomedical Research 2013; 24 (4): 509-514
Vascular endothelial growth factor (VEGF) plays a critical role in angiogenesis. Several VEGF polymorphisms were associated with development of different diseases in human. More commonly studied VEGF single nucleotide polymorphisms (SNPs) are -2578C/A, - 1154G/A, +936C/T. Our aim was to develop an asymmetric PCR on Light Cycler melting curve analyses for VEGF +936C/T to remove hook effect which is commonly observed in routine real-Time PCR with melting curve analyses. Real–time PCR with equimolar and asymmetric primer concentration and hybridization probes in LightCycler was performed and melting peaks for hook effects were compared. Melting curve was used for the genotyping. Asymmetric primer concentration (sense: antisense=3:1) neutralized the hook effect and resulted in the easy genotyping for the detection of VEGF SNP +936C>T. An asymmetric real-time PCR with melting curve analysis could be applied when hook effect is observed with common real-time PCR protocol.
Keywords
Asymmetric real-time PCR, hook effect, VEGF, melting curve analyses
Introduction
Vascular endothelial growth factor (VEGF) is a major angiogenic factor and a prime regulator of endothelial cell proliferation [1]. The VEGF gene is located at chromosome 6p21.3 and consists of eight exons. At least 30 single nucleotide polymorphisms (SNPs) in this gene have been detected [2]. Most commonly studied SNPs of VEGF are 460C/T, -1154G/A, -2578C/A, and +936C/T. A number of studies have shown an association of VEGF SNPs with different diseases such as breast cancer, prostate cancer [3-6], diabetic retinopathy [7], coronary arterial disease [8], Alzheimer’s disease [9], stroke [10], acute cerebral infarction [11], Parkinson's disease [12], and mi graine [13]. Our group is involved in studying the importance of VEGF SNPs (-2578C/A, -1154G/A, +936C/T) in strokes and associated diseases. Most of the studies used the PCR-RFLP technique for the determination of VEGF genetic polymorphism; which is very tedious and time consuming. A very few studies introduced Real-Time PCR followed by fluorescence–labeled probe hybridization and melting curve analysis by the Light Cycler [14-17]. However, with commonly used protocol for LightCycler real –time PCR, we experienced poor amplification and melting peaks in the VEGF SNP +936C/T with ‘hook effect’ phenomenon [18]. Compared with other optimizing parameters, the relative concentrations of sense primer and antisense primer are important and a few studies demonstrated the beneficial effect of asymmetric Real-time PCR. In this study, we carried out continuous analysis with various primer concentrations of an asymmetric Real-time PCR to nullify the hook effect and pro duce easily distinguishable melting peaks between wild type and mutated types.
Materials and Methods
After obtaining informed consent, peripheral blood samples were collected for genomic DNA isolation from stroke patients and controls attending the neurological clinic at CBNU, Jeonju, Korea. Stroke, a form of cerebrovascular disease, was defined as fast growing clinical sign of acute, focal or global disturbance of neurological function lasting more than 24 hours or symptoms lasting less than 24 hours and with evidence of a cerebral infarction in clinically relevant areas of the brain according to brain imaging either by CT or MRI and electrocardiography. DNA was extracted using Quick Gene DNA whole blood kit S (Life Science, Japan), according to manufacturer’s instruction. DNA purity and quantity were determined by absorbance reading at 260 and 280 nm in UV/Visible spectrophotometer, Amersham Bioscience, Cambridge, England. Real-time PCR with melting curve analysis were performed in the LightCycler (2.0) instrument (Roche Applied Science, Germany) for the detection of VEGF SNP +936C>T. The following primers and labeled probes were designed and synthesized by Sigma- Aldrich, Japan.
Forward primer: 5’-CACTTTGGGTCCGGAGGG-3’ Reverse primer: 5’-GGGCTCGGTGATTTAGCAG-3’ SensorProbe: 5’-AAGAGGGACCGTGCTGGGTCA [fluorescein]-3’ AnchorProbe: 5’-[LCRed640] CCGGGAATGCTTC CGCCGGAGTCT[Phos]-3’
Real-Time PCR was performed in glass capillary tubes (Roche Applied science). The PCR mixture of total volume 20 μl included 1.0-2.0 μl (5-10ng) of genomic DNA, 1.0μl of each primer (10.0 μM), 0.8 μl of each probe (10.0μM) and 4.0 μl of Light Cycler FastStart DNA Master HybProbe kit (Roche) which contains FastStart Taq DNA polymerase, reaction buffer, MgCl2 and dNTP mix and finally PCR grade water upto 20 μl. The cycling protocol consists of initial 15 min denaturation at 95°C followed by 45 cycles: denaturation (95 °C; 10 s) - annealing (59°C; 15s) - extension (72°C; 10s).
The temperature ramp rate was 20 °C/s. Emitted fluorescence was measured at the end of each annealing step at the F2/F1 channel. Immediately after amplification, melting curve analysis was performed on the LightCycler. The melting curve protocol included raising the temperature at 95 °C for 30 s, cooling to 45 °C for 1 min and slow heating to 85 °C at a rate of 0.2 °C/s. During this time fluorescence measurements were continuously collected in the F2/F1 channel. The plot of the fluorescence vs temperature (F2/F1 vs T) of the LightCycler (2.0) software is useful for the monitoring of a successful amplification and for the identification of different genetic alleles by their different melting peaks. But with this commonly used protocol for LightCycler Real–Time PCR, we experienced poor amplification and genotyping/melting peaks with ‘hook effect’ phenomenon for VEGF SNP +936C/T study. We performed the asymmetric Real-Time PCR in which we applied the different concentrations of sense and antisense primer (1:1, 1:3 and 3:1); as primers are more important among the standardization components and finally the ‘hook effect’ was nullified.
Results and Discussion
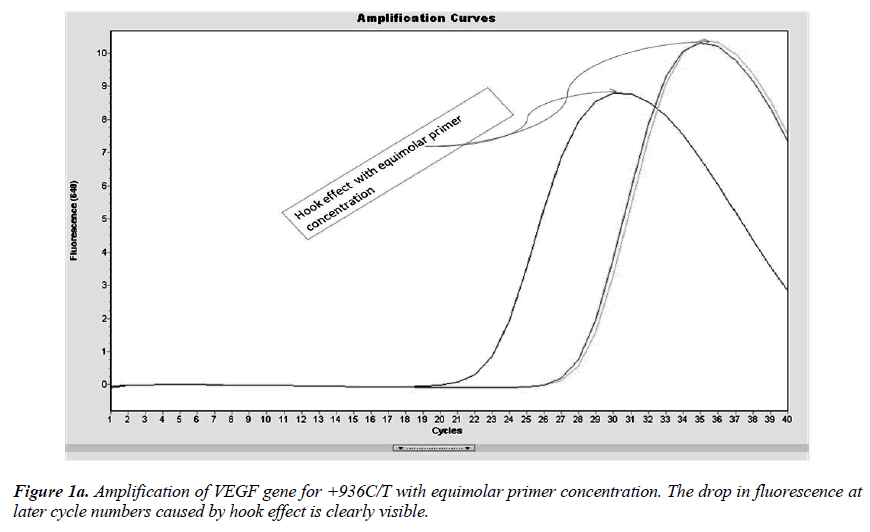
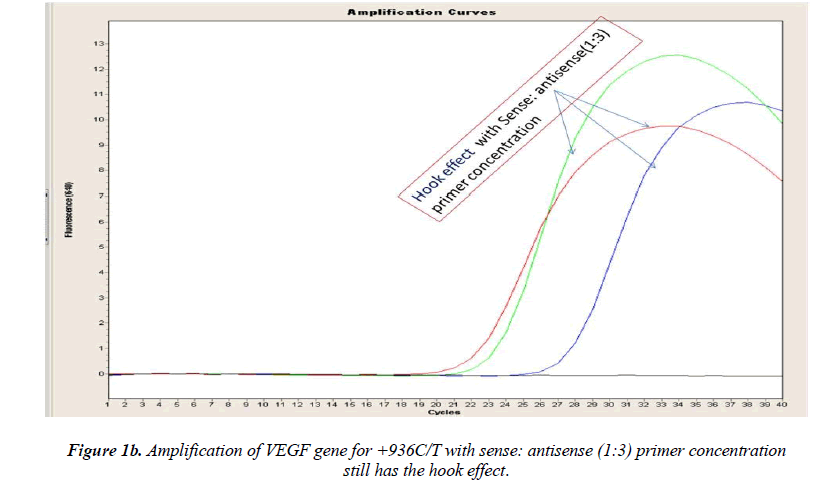
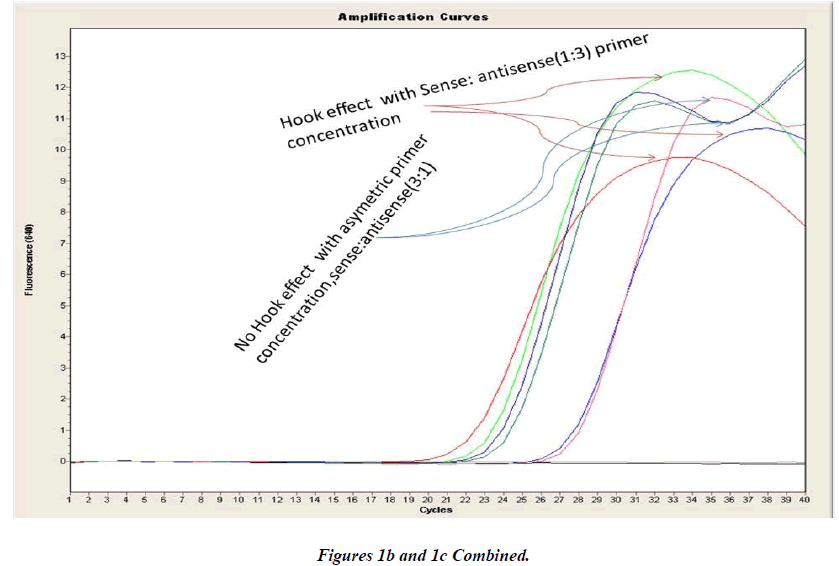
Real-time PCR amplification for the VEGF SNP +936C>T detection proved quite robust. The PCR product was free from other by-products or primer-dimers and at its correct size (119 bp) as estimated by agarose gel electrophoresis. By using common protocol of PCR (sense: antisense primer ratio= 1:1), we experienced ‘hook effect’ (Fig.1a and 1b), which is the sign of very efficient real- Time PCR [18]. In hook effect, the amplified DNA strands are reannealed before the probe can bind to generate fluorescence in later cycles of the PCR.
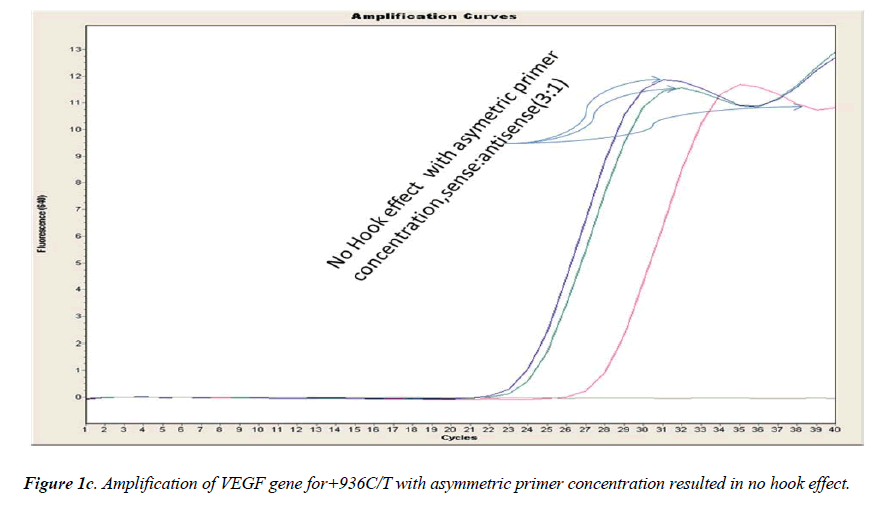
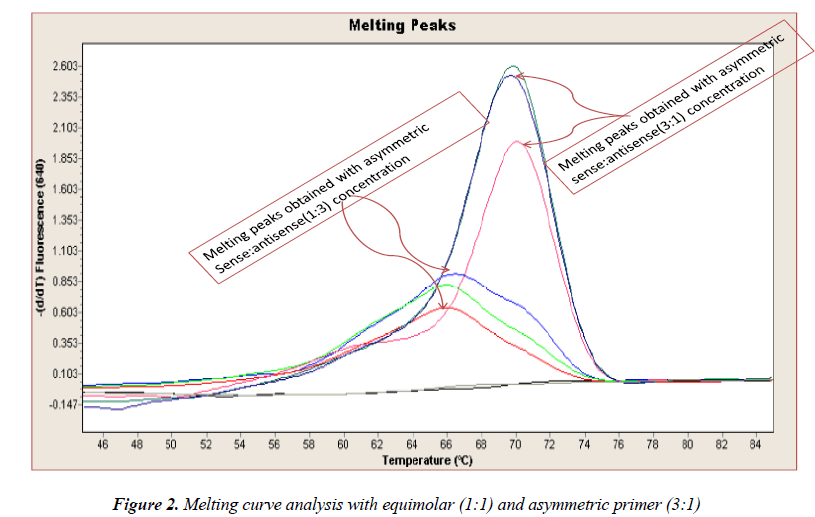
In general, it is not the problem of quantification where the crossing points (the cycle number at which the reaction enters exponential phase) are of interest but the SNP genotyping by melting curve analysis may be more problematic and we experienced the same. Among the standardization components, the relative amounts of primers are important and a few studies reported the advantages of asymmetric real time PCR (AS-RT-PCR) [15,19, and 20]. We resolved this problem by using asymmetric primer concentration in which we applied the different concentrations of sense vs antisense primers and the sense: antisense ratio (3:1) resulted in more of the strands complementary to the probe being amplified and more signal was obtained without ‘hook effect’ (Fig.1c). Thus, this intense optimization of asymmetric real-time PCR reactions resulted in the easy genotyping for the detection of VEGF SNP +936C>T and led to very good amplification efficiencies and a significant Tm difference between the peaks of normal and mutant alleles (Fig. 2).
Finally, the developed protocol is technically very simple and sound to perform and also there is less chance of contamination as there is no need of post-Amplification electrophoresis. Asymmetric real-time PCR followed by melting curve analysis is a rapid method (total time ~1 hour) for the detection of +936C>T mutation in VEGF. It is also a high throughput method since in the 32-position LightCycler carousel thirty samples plus a blank and a control heterozygote can be readily analyzed within an hour after DNA extraction and allocation of DNA to capillaries
In summary, an asymmetric real-time PCR with melting curve analysis could be applied for the screening of +936C>T mutation in VEGF. In the last but not the least, we would like to recommend the use of asymmetric primer concentration to prevent ‘hook effect’ and poor genotyping that results when conventional LightCycler genotyping is used.
Acknowledgment and source of funding
This work was supported by the research fund of the Biomedical Research Institute of Chonbuk National University Hospital.
References
- La Rosa S, Uccella S, Finzi G, Albarello L, Sessa F, Capella C. Localization of vascular endothelial growth factor and its receptors in digestive endo- crine tumors: correlation with microvessel density and clinicopathologic features. Hum Pathol 2003; 34: 18-27.
- Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vas- cular endothelial growth factor (VEGF) gene: corre- lation with variation in VEGF protein production. Cytokine 2000; 12: 1232-1235.
- McCarron SL, Edwards S, Evans PR, Gibbs R, Dearnaley DP, Dowe A,Southgate C, Easton DF,Eeles RA, Howell WM. Influence of cytokine gene polymorphisms on the development of prostate can- cer. Cancer Res 2002; 62: 3369-3372.
- Shahbazi M, Fryer AA, Pravica V, Brogan IJ, Ram-say HM, Hutchinson IV,HardenP N. Vascular endo- thelial growth factor gene polymorphisms are asso- ciated with acute renal allograft rejection. J Am Soc Nephrol 2002; 13: 260-264.
- Krippl P, Langsenlehner U, Renner W, Yazdani- Biuki B, Wolf G, Wascher TC, Paulweber B, Haas J, Samonigg H. A common 936 C/T gene polymor- phism of vascular endothelial growth factor is asso- ciated with decreased breast cancer risk. Int J Can- cer 2003; 106: 468-471.
- Lin JX, Tomimoto H, Akiguchi I, Matsuo A, Waki- ta H, Shibasaki H, Budka H. Vascular cell compo-nents of the medullary arteries in Binswanger’s dis- ease brains: a morphometric and immunoelectron microscopic study. Stroke 2000; 31: 1838-1842.
- Al-Kateb H, Mirea L, Xie X, Sun L, Liu M, Chen H,Bull SB, Boright AP, Paterson AD; DCCT/EDIC Research Group. Multiple variants in vascular en- dothelial growth factor (VEGFA) are risk factors for time to severe retinopathy in type 1 diabetes: the DCCT/EDIC genetics study. Diabetes 2007; 56: 2161-2168.
- Petrovic D. The role of vascular endothelial growth factor gene as the genetic marker of atherothrom-botic disorders and in the gene therapy of coronary artery disease. Cardiovasc Hematol Agents Med Chem 2010; 8: 47-54.
- Ignacio Mateo, Javier Llorca, Jon Infante, EloyRodr´ıguez-Rodr´ıguez, Coro S´anchez-Quintana a, Pascual S´anchez-Juan a,Jos´e Berciano, Onofre Combarros. Case-control study of vascular endothe- lial growth factor (VEGF) genetic variability in Alzheimer’s disease. Neuroscience Letters 2006; 401: 171-173.
- Kim OJ, Hong SH, Oh SH, Kim TG, Min KT, Oh D, Kim NK. Association between VEGF Polymor-phisms and Homocysteine Levels in Patients with Ischemic Stroke and Silent Brain Infarction. Stroke 2011; 42:2393-2402.
- Fu Y, Ni P, Ma J, Ying Y, Zhao J, Liu J, Chen S. Polymorphisms of human vascular endothelial growth factor gene are associated with acute cere- bral infarction in the Chinese population. Eur Neu- rol. 2011; 66: 47-52.
- Mihci E, Ozkaynak SS, Sallakci N, Kizilay F, Ya- vuzer U. VEGF polymorphisms and serum VEGF levels in Parkinson's disease. Neurosci Lett 2011; 494: 1-5.
- Gonclaves FM, Martins-Oliveira A, Speciali JG, Izidoro-Toledo TC, Luizon MR, Dach F, Tanus- Santos JE. Vascular endothelial growth factor ge- netic polymorphisms and haplotypes in women with migraine.DNA cell Biol 2010: 29: 357-362.
- Lyon E. Mutation detection using fluorescent hy- bridization probes and melting curve analysis. Ex- pert Rev Mol Diagn 2001; 1: 92-101.
- Lyondagger E, Millsondagger A, Phan T, Wittwer CT. Detection and identification of base alterations within the region of factor V Leiden by fluorescentmelting curves. Mol Diagn 1998; 3: 203-209.
- Nagy B, Savli H, Molvarec A, Várkonyi T, Rigó B, Hupuczi P, Rigó J Jr. Vascular endothelialgrowth factor (VEGF) polymorphisms in HELLP syndrome patients determined by quantitative real- time PCR and melting curve analyses. Clin Chim Acta 2008; 389: 126-131.
- Vural P, Küskü-Kiraz Z, Doğru-Abbasoğlu S, CilE, Karadağ B, Akgül C, Uysal M. Vascular endo- thelial growth factor -2578 A/C, -460 T/C and +405 G/C polymorphisms in polycystic ovary syndrome. Eur J Obstet Gynecol Reprod Biol 2009; 147: 57-60
- Technical note no. LC 8/99, Roche Molecular Bio- chemicals, 1999.
- Burggraf S, Kösel S, Lohmann S, Beck R, Olgemöller B. Unexplained DNA melting behavior in a ge- notyping assay. Clin Chem 2002; 48: 199-201.
- Barratt K, Mackay JF. Improving real-time PCRgenotyping assays by asymmetric amplification. J Clin Microbiol 2002; 40: 1571-1572.