ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 20
Aberrant SEPT9 methylation in plasma cell-free DNA of CRC patients
Mona Shayan1, Afsaneh Mojtabanezhad Shariatpanahi2, Seyed Morteza Seifati1 and Mohammad Amin Kerachian2,3,4*
1Medical Biotechnology Research Center, Ashkezar Branch, Islamic Azad University, Ashkezar, Yazd, Iran
2Cancer Genetics Research Unit, Reza Radiotherapy and Oncology Center, Mashhad, Iran
3Cancer Genetics Research Center, Mashhad University of Medical Sciences, Mashhad, Iran
4Department of Medical Genetics, Mashhad University of Medical Sciences, Mashhad, Iran
- *Corresponding Author:
- Mohammad Amin Kerachian
Cancer Genetics Research Unit
Reza Radiotherapy and Oncology Center
Mashhad, Iran
Accepted date: September 6, 2018
DOI: 10.4066/biomedicalresearch.29-18-951
Visit for more related articles at Biomedical ResearchColorectal cancer (CRC) is one of the most common cancers worldwide. CRC develops from precancerous polyps in the colon or rectum and is preventable by an early diagnosis and with the removal of precursor lesions. Numerous genetics and epigenetic alterations transform benign polyps to malignant tumors by affecting different pathways. Over the past decade, increasing evidence represent the utility of cell-free DNA as a ‘liquid biopsy’ to supplement non-invasive biopsies for genetic and epigenetic characterization and monitoring of solid cancers. One of the epigenetic biomarkers that has gained more attention in CRC is aberrant DNA methylation of Septin 9 gene. In this study we try to evaluate the methylation of Septin 9 gene status in the cfDNA of the plasma in colorectal cancer patients. Plasma cell-free DNA samples were extracted from 30 patients with background of tumors or polyps and 30 samples from healthy individuals. Septin 9 methylation analysis was performed by using the bisulfite specific high resolution melting analysis. The result showed a sensitivity and specificity of 10% and 53.33%, respectively. In conclusion, our results demonstrated that Septin 9 DNA methylation in plasma determined by THP and BSP-HRM had not have sufficient accuracy.
Keywords
Colorectal, Cancer, Methylation, SEPT9
Introduction
Colorectal cancer (CRC) is a malignancy that originates from the mucosal layer of the colon or rectum and adenomatous polyps [1]. CRC is the third and fourth most common cancer in men and women, respectively [2]. This causes 61,000 deaths annually around the world [3]. One of the most virulent tumors which has a second rank is CRC with an incidence of 13.1% in Europe and first ranked is belong to lung cancer [3]. A large number of patients are under the age of 50 years [4]. Screening of CRC is affordable cost which could be compared with other preventive method such as therapy of moderate hypertension [5]. CRC develops from precancerous polyps in the colon or rectum and is preventable and curable by an early diagnosis and with the removal of premalignant polyps. If tumor of CRC patients would be detected at early stage, the chance of survive will be increased. Adenomas as a premalignant lesion have a key role to diagnostic CRC [6]. Two approaches which are in used for CRC screening tests could be categorized: first on non-invasive tests for diagnosing primary cancer, such as guaiac fecal occult blood test (gFOBT), fecal immunochemical test (FIT) and stool DNA tests; second approach is invasive tests which could detect advanced lesions and cancer, such as double-contrast barium enema, colonoscopy, flexible sigmoidoscopy and virtual colonoscopy [7-9]. Colonoscopy tests cost a lot. Every 1,000 cases of colonoscopy, approximately one to five cases are associated with serious side effects [10]. In colonoscopy, tissue biopsy can be biopsied, which is a standard golden method for detecting colon cancer [11].
Today, various tissue analyses are done to diagnose and screen for CRC. One of these methods is genetic analysis [12]. Noninvasive biomarkers could be found in the stool or in plasma of patients and they are extremely sensitive and specific to evaluate genetic, epigenetic or protein markers [9,13-15]. Generally, the pathogenesis of CRC can be attributed to genetic and epigenetic changes in colon epithelium cells. Various mutations lead to genomic instability and play an important role in the development of CRC [16]. Chromosomal instability and micro-satellite molecular pathway involved in the development of CRC [17]. The methylation of genes as an epigenetic process could have a potential role in colorectal carcinogenesis [16-19]. One of the epigenetic biomarkers that has gained more attention in CRC is aberrant DNA methylation of Septin 9 (SEPT9) gene. Methylated SEPT9 could be detected on cell-free tumor DNA. Almost every report on circulating DNA identifies apoptosis or necrosis or both as the main source of free circulating tumor DNA in serum and plasma [20-23]. Septin 9 proteins belong to upper class of P-loop GTPases and they are a group of GTPbinding proteins. Also, one of the main factors of cell division in yeast is Septin genes [24]. SEPT9 has a major task in many cellular processes, like providing strength to the cell wall, recruiting proteins to specific subcellular locals to serve as scaffolds, making membrane diffusion barriers to create separate cellular domains and they have a role in cell polarity determination [24,25]. The molecular functionality of Septin 9 (SEPT9) is not still discovered tumorigenesis of colon; the mentioned gene contains 18 unique transcripts which is encodes 15 polypeptides and generated by alternative splicing and its research has not been completed [26]. Methylated SEPT9 (mSEPT9) has found in CRC cases and patients with precancerous lesions such as adenomas [21-25,27]. The objective of this research was to study aberrant DNA methylation of SEPT9 gene in plasma of patients with pioneer lesions of CRC.
Materials and Methods
Study participants
This was a case-control study. Patients with sporadic CRC who participated in this study were recruited consecutively from April 2015 to March 2017. CRC tissues were collected during colonoscopy from 60 patients referred to Reza Radiotherapy and Oncology Center (RROC, Mashhad, Iran). In total, 30 polyp/tumor positive patients and 30 patients with normal colons diagnosed by colonoscopy were enrolled in this study. Histopathology reports were assessed to determine polyp/ tumor characteristics. Patients with prior colorectal resection and history of any cancer or chemotherapy or radiation therapy were excluded from this study. In order to reduce bias, we designed this experiment as a blinded assay and samples were randomly coded before processing. All sample collection and preservation were taken care of by an individual who did not participate in the follow-up studies. All patients gave informed written consent to participate and to have their biologic specimens analysed. The study was approved by the Ethical Committee Mashhad University of Medical Sciences, Iran.
Collection of plasma
Five ml peripheral blood was collected from patients and healthy individuals into EDTA tubes and kept at room temperature (18-22°C). Plasma was separated by double centrifugation (800 g; 10 min, separation, 1600Xg; 10 min), no more than 2 h after blood draw. Plasma aliquots were immediately frozen at -70 °C.
Cell-free DNA extraction (cfDNA)
cfDNA purification was performed by the standard Triton/ Heat/Phenol protocol (THP) method, which removes proteins from nucleic acids by mixture of phenol-chloroform-isoamyl alcohol. Briefly, in this method 500 μl of plasma was mixed with 5 μl Triton X-100 (Applichem, Germany) and heat denatured at 98°C for 5 min. Samples were placed on ice for 5 min, then extracted with an equal volume of phenolchloroform- isoamyl alcohol (25:24:1, v:v:v), saturated with 50 Mm tris-Cl, pH 8.0 and centrifuged for 10 min at 14,000Xg. The aqueous phase was precipitated for 2 h with × 2.5 volume of 100% ethanol at -70°C. The DNA pellet was washed with 1 ml ethanol 70%, air-dried and re-suspended in 50 μl of AE buffer (10 mM tris-Cl, 0.5 mM EDTA; pH 9.0) and incubated overnight at 37°C.
Bisulfite treatment
Twenty μl extracted cfDNA undergone sodium bisulfite conversion and DNA recovery using the EpiTect Fast Bisulfite Conversion Kits (Qiagen, Germany) according to the manufacturer’s instructions.
Methylation analysis
Methylation analysis was performed by bisulfite specific high resolution melting analysis (BS-HRM) consisting of PCR amplification of bisulfite-modified DNA. The primers used to amplify bisulfite-treated DNA were SEPT9-F 5'- TTTATTTAGTTGAGTTAGGGGGTTTA-3' and SEPT9-R 5' AACCCAACACCCACCTTC-3', designed to amplify both methylated and unmethylated bisulfite-treated DNA that did not amplify unmodified genomic DNA.
PCR amplification and HRM analysis were carried out sequentially on a light Cycler® 96 System (Roche, Germany). PCR was carried out in a 10 μl total volume using HiFiSYBR Green Master Mix (Farabin, Tehran), consisting of 2.5 μl of bisulfite modified template, 0.2 μg/μl BSA and 300 nM of each primer. The amplification run was 15 min at 95°C, followed by 45 cycles of 20 s 95°C, 15 s at the primer annealing temperature (60°C) and 15 s at 72°C. HRM analyses were performed at the temperature ramping from 65 to 97°C. Florescence acquisition setting was carried out at temperature recommended by the manufacturer. The melting curves were normalized by calculation of the ‘line of best fit’ in between two normalization regions before and after the major fluorescence decrease representing the melting of the PCR product using the software version 1.1 provided with the LightCycler® 96 System.
Statistical analysis
The sensitivity and specificity (with 95% confidence interval (CI)) of the Septin 9 hypermethylation of cfDNA plasma were calculated. To compare characteristics of the different groups of patients and samples, t-test for continue variables, Chisquare test and Fisher exact test were used for categorical variables. Statistical analyses were performed using SPSS version 13.0. All values were two-sided and P value<0.05 was considered to indicate a statistically significant difference.
Results
Patient and lesion characteristics
The clinical characteristics of the 60 patients included in this study were shown in Table 1. There was no significant difference with respect to gender and bone mass index (BMI) between cases and controls (p value>0.05). The BMI of individuals with polyps was 24.99%, which was not significantly different from the bodyweight of healthy individuals, which was 25%. In this research, patients with polyps had a weight loss of 20%. 16.66% of those were close to their ideal weight, 40% overweight, and 23.33% remained very obese.
| Characteristics | Polyp/tumor | |
|---|---|---|
| Negative | Positive | |
| Sex | ||
| Female | 13 (43.33%) | 12 (40%) |
| Male | 17 (56.66%) | 18 (60%) |
| Age group (y) | 50.56 (15-79) | 58.66 (30-76) |
| Body mass index (kg/m2) | ||
| Underweight (BMI<18.5) | 0 | 6 (20%) |
| Healthy weight (BMI: 18.5-24.9) | 9 (30%) | 5 (16.66%) |
| Over weight (BMI: 25-29.9) | 12 (40%) | 12 (40%) |
| Obese (BMI of 30 or greater) | 9 (30%) | 7 (23.33%) |
| Hx. of drug intake | ||
| Yes | 1 (3.33%) | 3 (10%) |
| No | 29 (96.66%) | 27 (90%) |
| Hx. of smoking | ||
| Yes | 8 (26.66%) | 9 (30%) |
| No | 30 (100%) | 29 (96.66%) |
| Location | ||
| Ascending colon | 7 (18.91%) | 4 (10.81%) |
| Rectum | - | 7 (18.91%) |
| Sigmoid | - | 17 (45.94%) |
| Transvers colon | - | 1 (2.7%) |
| Descending colon | - | 5 (13.51%) |
| Cecum | - | 3 (8.1%) |
| Results of pathology | - | |
| Tubular adenoma | - | 17 (56.66%) |
| Tubulovillous adenoma | - | 5 (16.66%) |
| Vilous adenoma | - | 0 (0%) |
| Hyperplastic polyp | - | 3 (10%) |
| High grade adenoma | - | 0 (0%) |
| Adenocarcinoma | - | 5 (16.66%) |
| Adenoma size ≥ 1 cm | - | 18 (60%) |
Table 1. Patient and lesion characteristics.
SEPT9 methylation status
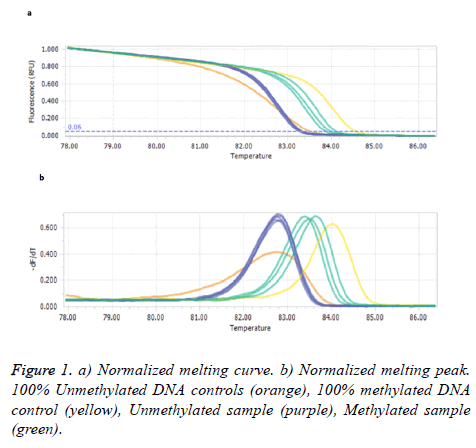
Figure 1 illustrates the comparison of the melting profiles of PCR products from samples with profiles specific for PCR products derived from methylated and unmethylated control DNAs.
Our results showed methylated SEPT9 test had a sensitivity and specificity of 10% and 53.4% in patients’ plasma with polyps/tumor, respectively. Statistical test analysis revealed that SEPT9 methylation in plasma was not significantly different in patients with control groups (P>0.05) as shown in Table 2.
| Characteristics/Polyp/Tumor | Positive (Methylated) | Negative (Un-methylated) | Sensitivity | Specificity | P-value |
|---|---|---|---|---|---|
| Positive | 3 | 27 | 10% | 53.33% | 0.7 |
| Negative | 14 | 16 |
Table 2. The performance of SEPT9 methylation test in plasma samples of CRC patients.
Discussion
In this study, we aimed to assess the potential role of aberrant SEPT9 promoter methylation changes in cfDNA released by tumor cells in different forms and at different levels in the blood circulation of CRC patients. We demonstrated that there was not significantly a higher frequency (P value>0.05) of SEPT9 methylated DNA in plasma of patients with polyps/ tumor versus healthy individuals with a sensitivity and specificity of 10 and 53.4%, respectively.
According to several comprehensive screening researches, patients have early screening test, would have more chances for surviving compared to those who did not undergo any screening test [28-31]. A study performed in 2014, proved that most of the patients (83%) were willing to accept mSEPT9 examine which is higher than colonoscopy (37%) and finally stool test (15%) [32]. According to studies conducted by Muller et al. a high degree of SEPT9 sensitivity in plasma has made it a better way to detect CRC than FBOT and CEA [33]. Song et al. accomplished a SEPT9 gene methylation test to diagnose CRC. A huge variety in sensitivity from 48.2% to 95.6% was detected as well as specificity from 100% to 80% [34]. In a study by Fu et al. sensitivity and specificity of 61.22% and 93.7%, for methylated SEPT9 (mSEPT9) in plasma CRC cases, was reported respectively. They also displayed that plasma mSEPT9 in monitoring CRC recurrences or metastases were reliable marker [35]. Besides, Wu el al. validated a simplified SEPT9 gene methylation assay in 1031 subjects in Chinese patients. The sensitivity and specificity for CRC detection was 76.6% and 95.9%, respectively. Their results indicated a satisfactory detection rate for all stages of CRC, including early stages [36]. Toth et al. assessed SEPT9 methylation in both tissue and plasma of healthy individuals, adenoma and CRC patients, and detected the methylated gene in all tissue samples at different levels regardless of the type. They realized that methylated SEPT9 levels in CRC and adenoma tissue samples were not significantly different; however, its levels in adenoma or CRC cases were much higher and considerably distinct from healthy tissue samples [37]. Previously in a study by Lee et al. they showed that fecal immunochemical test also exhibited a high sensitivity for colon cancer similar to plasma SEPT9 methylation [38]. The sensitivity of plasma mSEPT9 for CRC was consistent with the data in a quantitative meta-analysis by Zhang et al., which showed that plasma methylated SEPT9 had a sensitivity of 64% (95% CI: 59%-68%) for CRC detection in the Asianbased population [39]. Epi proColon® 2.0 CE is based on methylated SEPT9 gene from the cfDNA in the plasma which is accessible in Europe and different nations such as china [40,41].
The low sensitivity and specificity in cfDNA-based studies could be due to different reasons. First, due to the very fragmented and low concentration of cfDNA in plasma, cfDNA extraction method plays a critical role. Some extraction methods such as THP generate a lower quality and quantity of cfDNA. Second, the specificity of SEPT9 methylation in plasma is influenced by the background normal SEPT9 methylation status. Aberrant cfDNA can be mixed by normal cell free DNA, shedded in the bloodstream (one percent of total cfDNA). cfDNA in the blood of cancer patients is not only representative of tumor derived DNA, but also of DNA released by healthy cells under different conditions [42,43]. Third, intertumoral heterogeneity could result in more complexity [44]. Therefore, these reasons may cause false positive/negative results.
This research demonstrates a sensitivity and specificity of 10 and 53.33%, respectively. Compare to other investigations, in the current study having several potential heteroduplexes generated by heterogeneous methylated CpG-rich amplicons is a challenge in BSP-HRM. It is hard to compare the homogenous methylated and unmethylated controls with melting HRM profile of heterogeneous methylated DNA samples [15]. Besides, the THP extraction method used in the current study could exacerbate the results and underestimate the sensitivity and specificity of SEPT9 methylation in plasma.
In conclusion, our results demonstrated that SEPT9 DNA methylation in plasma determined by THP and BSP-HRM had not have sufficient accuracy.
References
- Williams CB, Saunders BP, Talbot IC. Endoscopic management of polypoid early colon cancer. World J Surg 2000; 24: 1047-1051.
- Hessami Arani S, Kerachian MA. Rising rates of colorectal cancer among younger Iranians: is diet to blame? Current oncology (Toronto, Ont) 2017; 24: 131-137.
- Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P. Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol Off J Eur Soc Med Oncol 2007; 18: 581-592.
- https: //www.nih.gov/about-nih/what-we-do/nih-almanac/national-cancer-institute-nci.
- Pignone M, Saha S, Hoerger T, Mandelblatt J. Cost-effectiveness analyses of colorectal cancer screening: a systematic review for the U.S. Preventive Services Task Force. Ann Int Med 2002; 137: 96-104.
- Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M. Genetic alterations during colorectal-tumor development. N Engl J Med 1988; 319: 525-532.
- Smith RA, Cokkinides V, Brooks D, Saslow D, Brawley OW. Cancer screening in the United States, 2010: a review of current American Cancer Society guidelines and issues in cancer screening. Cancer J Clinic 2010; 60: 99-119.
- Kormi SMA, Ardehkhani S, Kerachian MA. New insights into colorectal cancer screening and early detection tests. Colorect Cancer 2017; 6: 63-68.
- Mojtabanezhad Shariatpanahi A, Yassi M, Nouraie M, Sahebkar A, Varshoee Tabrizi F, Kerachian MA. The importance of stool DNA methylation in colorectal cancer diagnosis: A meta-analysis. PloS One 2018; 13: 0200735.
- Delavari A, Bishehsari F, Salimzadeh H, Khosravi P, Delavari F, Nasseri-Moghaddam S. Adenoma detection rates in an opportunistic screening colonoscopy program in Iran, a country with rising colorectal cancer incidence. BMC Gastroenterol 2014; 14: 196.
- Schoen RE, Pinsky PF, Weissfeld JL, Yokochi LA, Church T, Laiyemo AO. Colorectal-cancer incidence and mortality with screening flexible sigmoidoscopy. N Engl J Med 2012; 366: 2345-2357.
- Ahlquist DA TW, Yab TC, Devens ME, Mahoney DW. Aberrantly methylated gene marker levels in stool: effects of demographic, exposure, body mass, and other patient characteristics. J Mol Biomark Diagn 2012; 3: 133.
- Kim HJ, Yu MH, Kim H, Byun J, Lee C. Noninvasive molecular biomarkers for the detection of colorectal cancer. BMB Rep 2008; 41: 685-692.
- Lange CP, Campan M, Hinoue T, Schmitz RF, van der Meulen-de Jong AE, Slingerland H. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PloS One 2012; 7: 50266.
- Rokni P, Shariatpanahi AM, Sakhinia E, Kerachian MA. BMP3 promoter hypermethylation in plasma-derived cell-free DNA in colorectal cancer patients. Gene Genom 2018; 40: 423-428.
- Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 2008; 135: 1079-1099.
- Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Pathol Res Int 2011; 2011: 902674.
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature 1998; 396: 643-649.
- Barati Bagerabad M, Tavakolian S, Abbaszadegan MR, Kerachian MA. Promoter hypermethylation of the eyes absent 4 gene is a tumor-specific epigenetic biomarker in Iranian colorectal cancer patients. Acta Medica Iranica 2018; 56: 21-27.
- van der Vaart M, Pretorius PJ. The origin of circulating free DNA. Clin Chem 2007; 53: 2215.
- Warren JD, Xiong W, Bunker AM, Vaughn CP, Furtado LV, Roberts WL, Fang JC, Samowitz WS, Heichman KA. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med 2011; 9: 133.
- deVos T, Tetzner R, Model F, Weiss G, Schuster M, Distler J. Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem 2009; 55: 1337-1346.
- Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 2008; 54: 414-423.
- Hall PA, Russell SE. The pathobiology of the septin gene family. J Pathol 2004; 204: 489-505.
- Estey MP, Kim MS, Trimble WS. Septins. Curr Biol 2011; 21: R384-387.
- McDade SS, Hall PA, Russell SE. Translational control of SEPT9 isoforms is perturbed in disease. Human Mol Gene 2007; 16: 742-752.
- Tanzer M, Balluff B, Distler J, Hale K, Leodolter A, Rocken C. Performance of epigenetic markers SEPT9 and ALX4 in plasma for detection of colorectal precancerous lesions. PloS One 2010; 5: 9061.
- Hardcastle JD, Chamberlain JO, Robinson MH, Moss SM, Amar SS, Balfour TW. Randomised controlled trial of faecal-occult-blood screening for colorectal cancer. Lancet (London, England) 1996; 348: 1472-1477.
- Kronborg O, Fenger C, Olsen J, Jorgensen OD, Sondergaard O. Randomised study of screening for colorectal cancer with faecal-occult-blood test. Lancet (London, England) 1996; 348: 1467-1471.
- Lindholm E, Brevinge H, Haglind E. Survival benefit in a randomized clinical trial of faecal occult blood screening for colorectal cancer. Br J Surg 2008; 95: 1029-1036.
- Mandel JS, Church TR, Ederer F, Bond JH. Colorectal cancer mortality: effectiveness of biennial screening for fecal occult blood. J Nat Cancer Inst 1999; 91: 434-437.
- Adler A, Geiger S, Keil A, Bias H, Schatz P, deVos T. Improving compliance to colorectal cancer screening using blood and stool based tests in patients refusing screening colonoscopy in Germany. BMC Gastroenterol 2014; 14: 183.
- Muller AD, Sonnenberg A. Prevention of colorectal cancer by flexible endoscopy and polypectomy. A case-control study of 32,702 veterans. Ann Int Med 1995; 123: 904-910.
- Song L. LYMSGiaSBfaSoCCCC 1: 1.
- Fu B, Yan P, Zhang S, Lu Y, Pan L, Tang W. Cell-free circulating methylated sept9 for noninvasive diagnosis and monitoring of colorectal cancer. Dis Mark 2018; 2018: 6437104.
- Wu D, Zhou G, Jin P, Zhu J, Li S, Wu Q. Detection of colorectal cancer using a simplified sept9 gene methylation assay is a reliable method for opportunistic screening. J Mol Diagn 2016; 18: 535-545.
- Toth K, Wasserkort R, Sipos F, Kalmar A, Wichmann B, Leiszter K. Detection of methylated septin 9 in tissue and plasma of colorectal patients with neoplasia and the relationship to the amount of circulating cell-free DNA. PloS One 2014; 9: 115415.
- Lee JK, Liles EG, Bent S, Levin TR, Corley DA. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. Ann Int Med 2014; 160: 171.
- Zhang M, He Y, Zhang X, Zhang M, Kong L. A pooled analysis of the diagnostic efficacy of plasmic methylated septin-9 as a novel biomarker for colorectal cancer. Biomed Rep 2017; 7: 353-360.
- Jin P, Kang Q, Wang X, Yang L, Yu Y, Li N. Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol 2015; 30: 830-833.
- Lamb YN, Dhillon S. Epi proColon((R)) 2.0 CE: a blood-based screening test for colorectal cancer. Mol Diagn Ther 2017; 21: 225-232.
- Danese E, Minicozzi AM, Benati M, Montagnana M, Paviati E, Salvagno GL. Comparison of genetic and epigenetic alterations of primary tumors and matched plasma samples in patients with colorectal cancer. PloS One 2015; 10: 0126417.
- Diaz LA, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol Off J Am Soc Clin Oncol 2014; 32: 579-586.
- Kamat AA, Bischoff FZ, Dang D, Baldwin MF, Han LY, Lin YG. Circulating cell-free DNA: a novel biomarker for response to therapy in ovarian carcinoma. Cancer Biol Ther 2006; 5: 1369-1374.