ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Computational Life Sciences and Smarter Technological Advancement: Edition: II
A voyage on medical image segmentation algorithms
Kumar SN1, Lenin Fred A2*, Muthukumar S3*, Ajay Kumar H4 and Sebastian Varghese P5
1Department of ECE, Sathyabama University, Jeppiaar Nagar, Rajiv Gandhi Salai, Chennai, India
2School of CSE, Mar Ephraem College of Engineering and Technology, Elavuvilai, Tamil Nadu, India
3Department of IT, Indian Institute of Information Technology, Srirangam, Trichirappalli, Tamil Nadu, India
4School of ECE, Mar Ephraem College of Engineering and Technology, Elavuvilai, Tamil Nadu, India
5Consultant Radiologist, Metro Scans and Laboratory, Thiruvananthapuram, Kerala, India
- *Corresponding Author:
- Lenin Fred A
School of CSE
Mar Ephraem College of Engineering and Technology
Tamil Nadu, India
Muthukumar S
Department of IT
Indian Institute of Information Technology
Tamil Nadu, India
Accepted on March 22, 2017
DOI: 10.4066/biomedicalresearch.29-16-1785
Visit for more related articles at Biomedical ResearchImage segmentation is the process of subdividing an image into eloquent regions that are consistent and homogeneous in some characteristics. Image segmentation is indeed a vital process in the early diagnosis of abnormalities and treatment planning. The segmentation algorithms are employed to extract the anatomical structures and anomalies from medical images. The segmentation algorithms can be categorized into three generations. The first generation algorithms are based on threshold, seed point selection and edge tracing methods. The second generation algorithms incorporate uncertainty and optimization models and the third generation algorithms considers the prior information in segmentation process. This review work discusses and conceptualizes the various segmentation algorithms, which are in correlation with medical images and adduce the result of some of the significant algorithms in each generation. Moreover, the proposed work does spell out the pros and cons of the algorithms for computer aided analysis. In extension, this literature review indeed paves an ample platform to the researchers for better understanding of various segmentation techniques and its characteristics for medical images.
Keywords
Segmentation, Preprocessing, Thresholding, Deformable models, Clustering.
Introduction
Medical imaging is a technique used to generate images of the human body for clinical purpose. Medical imaging techniques have revolutionized the modern medical era. Medical imaging techniques such as Computer Tomography (CT), Magnetic Resonance Imaging (MRI) and Ultrasound (US) are widely used for the diagnosis of anomalies like cyst, tumor etc. CT can generate images at much higher spatial resolution with shorter imaging duration [1]. MRI and US takes up the advantage of not exposing the subject to ionizing radiation [1]. Image segmentation is the process of dissection up of an image into useful parts, often consisting of an object and background. Despite of the choice of ‘n’ number of segmentation algorithms, selection is based on the application and image characteristics. In medical imaging context, segmentation is a segregating operation of an image domain into nonoverlapping sets of pixels regions that corresponds to notable anatomical structures or anomalies such as tumor or cyst. The segmentation of anatomical organs plays a vital role in many diagnostic and surgical procedures and it is also useful in computer aided diagnosis, computer guided surgery systems and building anatomical atlases. The segmentation algorithms can be widely categorized into three types viz, supervised, unsupervised and interactive. Supervised segmentation methods need manually labeled training data for recognizing specific region of interest in images, which may restrict the scope of this method [1-4]. Unsupervised (automatic) methods provide segmentation results without prior information about the input images and don’t require manual intervention [5]. The unsupervised segmentation methods generally impose limits on the segmentation of complex region of interest [6,7]. In interactive segmentation the accuracy, precision and efficiency was enhanced by the combination of human experts and machine intelligence [4]. The interactive segmentation takes a prominent role for various applications like localization of tumors, estimating tissue volumes, computer aided surgery, and diagnosing diseases [8]. The features are extracted from the segmented region of interest for classification and recognition (e.g. classifications in medical domain are the conditions like normal, benign, malignant) [9]. Some of the widely used segmentation algorithms with comparative analysis which includes the results with special concentration in medical image processing are also discussed in this paper.
Segmentation Algorithms
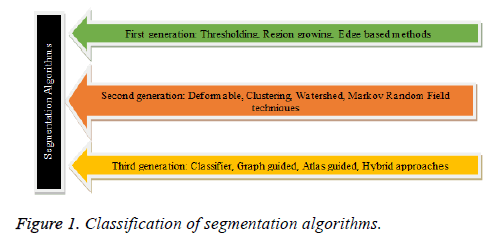
The medical images retrieved from the acquisition system, are first subjected to suitable preprocessing technique to minimize the noise, artifacts and bias effects. The preprocessing plays a vital role and its prime objective is to convert the input image in an apt format for subsequent process like segmentation, feature extraction and classification. The CT and MR images comprises of partial volume effects, artifacts and noise due to sensors and electronic system [10,11]. The CT images in general are perverted by Gaussian noise and artifacts, MR images are distorted by rician noise, artifacts , intensity inhomogeneity due to the non-uniform response of RF coil and the ultrasound images are corrupted by speckle noise and artifacts [10,12]. The intensity inhomogeneity of MR images can be contemplated as the spatially varying bias field multiplied by the true signal under measurement and the bias correction involves the estimation of bias field and correction of intensity inhomogeneity [10]. A wide number of spatial and transform domain algorithms are there for preprocessing and the choice of the algorithm depends on the image modality and the noise characteristics [10-13]. The various techniques for intensity inhomogeneity correction are surface fitting method (based on intensity or gradient), histogram based methods, high frequency maximization methods, filtering methods (homomorphic filtering) [14,15]. A wide variety of segmentation algorithms are indeed accessible for the extraction of desired region of interest in medical images. The segmentation algorithms are classified based on its characteristics, time of evolution and are depicted in Figure 1.
First Generation Segmentation Algorithms
Thresholding is the basic segmentation algorithm to create binary images in which the pixels having gray level intensity greater than the threshold are in the foreground region and the pixels having threshold value less than the threshold are in the background region [16]. The thresholding approach works fine for high contrast objects with a sharp edge, but it often fails as soon as the edges are smooth of varying intensity, influenced by noise and blur in boundaries [17]. The thresholding algorithm is a low level segmentation technique and relies on the local spatial domain characteristics. The thresholding techniques are strenuous and time consuming in volumetric data set. Region based segmentation algorithm comprises of mainly the region growing, region splitting and region merging techniques. In region growing algorithm, based on the similarity criteria of the seed point with the neighboring pixels, the growing of region will be done and the similarity criteria can be gray level intensity, shape, size or color etc. [18]. The region merging algorithm merges the regions that are homogeneous based on the predefined similarity criteria. The selection of seed point and the similarity criterion are vital factors that reflect the segmentation result of region growing technique [18]. The edges are significant in many applications and it is basically a convolution operation done on an image by a mask with appropriate values. The canny is an efficient edge detector that encompasses of edge enhancement and edge tracing stage [19]. In the case of noisy input images, edge detection has to be preceded by a restoration stage for smoothing the image while preserving the edges. The smoothing of the image may sometimes leads to false edge detection and can be eliminated by multiresolution edge detection and edge tracing techniques [20]. The region merging after region growing eliminates the high frequency artifacts with seed point selection based on the local statistics and was used for the analysis of breast mass and liver cyst [18]. The region growing algorithm with edge detection and morphological operations segments the lung parenchyma on CT images for diagnosing lung diseases [21]. An adaptive region growing algorithm based on the mean of intensity values and norm of the intensity gradient in the neighborhood region of the pixels was developed for the segmentation of bones in 3D [18F] fluoride ion PET images [22]. A region merging algorithm was proposed based on the Euclidean distance as the similarity measure for the segmentation of bones in radiographic images of hand taking in to account of the similarity constraints in local and extended region [23]. In [24] the fuzzy information was incorporated in the conventional region growing seed point selection for the segmentation of tissues in MR brain images. Samy et al. [25] affirmed that the fuzzy based tsallis entropy Thresholding produces satisfactory results for MR brain and abdomen CT images in the presence of noise and image with complex backgrounds. Sheema et al. proposed the multilevel thresholding based on Shannon, Renyi and Tsallis entropy for the segmentation of skin lesions; the Shannon and Renyi entropy approach produces good results [26]. In [27], the authors proposed Multiscale Region Growing (MSRG) technique for coronary artery segmentation in 2D X-ray angiograms. The MSRG with Frangi filter produces good results and is insensitive to noise and artifacts.
Second Generation Segmentation Algorithms
Deformable models-energy minimization based approach
In active contours model, an initial contour comprising of a set of discrete points is placed on the boundary of desired region of interest. The evolution of the curve takes place with respect to a set of constraints. The classical snake model comprises of internal energy (elasticity and rigidity term) and external energy (image term and user term). The elastic energy term will make the snake to act like membrane and the rigidity term determines the bending energy of the contour to behave as a thin plate. The image term aids the contour in finding the desired region of interest. The user term helps the snake model to eliminate the problem in the framing of initial contour and to make the snake model insensitive to noise generated during the acquisition of images [28]. In classical snake algorithm, positioning of discrete set of points should be close to the desired region of interest, hence much user intervention is needed and it is also sensitive to noise having high noise to signal ratio [29,30]. Some of the variants of classical snake model for preserving topology are greedy snake model, Tsnakes and T-surfaces, balloon model, gradient vector flow and dynamic programming spline. The dynamic programming spline is a parametric model in which the deformable model is split into parts by knot points [31]. Each curve is segmented as piecewise polynomial function and this model is sensitive to topological changes during the evolution of curve [31]. The balloon model encompasses a pressure force component to the external energy term and the expansion or contraction of contour occurs with constant speed [32]. The level set algorithm is a curve evolution technique in which the evolution of the contour is the zero level set of the higher dimension function [33]. The main feature of level set algorithm is its ability to change the topology easily, however sometimes during the curve evolution, splitting or vanishing of the contour can occur leading to inefficient result. The geodesic active contour is a geometric deformable model which adapts the features from level set and snake algorithm [34]. The expression for energy minimization in geodesic active contour comprises of attraction component and regularity component (rigidity constraints are set to zero) [35]. Chan Vese deformable model proposed the stopping criterion for curve evolution based on Mumford Shah model and thereby it minimizes the computational complexity in level set algorithm [36]. Active contour model with Principal Component Analysis (PCA) for training curve shape was capable of segmenting cardiac and prostate glands in MR images and is insensitive to noise [37]. The levels set model with bias correction for intensity in homogeneities was framed for segmentation of tumor mass in breast MR images [38]. A Gaussian Regularizing Level Set Method (GRLSM) with Local Image Fitting (LIF) algorithm was used for the segmentation of brain tumor in MR images [39]. Zhou et al. considered a region based active contour model comprising of external energy, internal energy and regularization term for the segmentation in synthetic and real medical images [40]. The shape prior was incorporated in the level set model for the segmentation of corpus callosum from 2D MR images and liver from 3D CT volumes [41]. The conventional level set model is susceptible to noise and weak boundaries, in some cases the region based level set do not give satisfactory results. A novel level set model based on fuzzy clustering was proposed for the segmentation of liver tumor on CT images that produces efficient result and it can be termed as a hybrid segmentation model [42]. The adaptive bias field regularization prior to level set model produces efficient segmentation result for synthetic and MR medical images with intensity inhomogeneity [43].
Watershed approach
In watershed segmentation algorithm the gray scale image is visualized in the form of topographical surface [44]. When a drop of water fall on a surface it will trace the path towards local minimum of the terrain and the catchment basin is collection of such points comprising of water droplets falling on the path of local minima. The pixels in the gray scale image are analogous to water droplets and the resultant catchment basin forms the sub region of the image having similar group of pixels. The watershed lines are the set of discrete points that separate catchment basins [45]. The watershed segmentation algorithm based on chessboard distance can yield good results than algorithms based on Euclidean distance and city block distance [46]. The watershed-flooding algorithm is faster than watershed rainfall algorithm, but it is not applicable for the segmentation of images with weak boundaries. The conventional watershed algorithms based on gradient and morphological operations are susceptible to over segmentation [47]. The issue of over segmentation in conventional watershed algorithm can be minimized by suitable preprocessing and post-processing techniques [47]. The over segmentation can be suppressed by choosing appropriate filtering technique (such as median filter, anisotropic diffusion filter) there by eliminating the irrelevant local minima [5]. The power watershed algorithm solves the issues in conventional watershed algorithm thereby producing the optimal result for the segmentation of multiple objects [48]. Marker controlled watershed algorithm based on morphological operations was used for the segmentation of tumor in MR images of brain and is insensitive to noise and over segmentation is eliminated [49]. The internal marker is associated with the ROI and the external marker is associated with the background [50]. An interactive watershed algorithm based on morphological operators and markers was developed for the segmentation of brain, cardiac ventricle on MR images [51]. The watershed segmentation always produce closed contour of the region of interest and watershed models based on wavelet, markers, scale space approach (based on partial differential equation) provides better result than conventional watershed approach [50,51]. The accuracy of the extracted contour in marker controlled watershed algorithm was improved by stochastic watershed algorithm. [52]. Zhe et al. applied watershed algorithm with contourlet transform for the segmentation of prostate in MR images. The preprocessing of the images was performed by contourlet transform and the texture gradient was combined with the marker controlled watershed algorithm that minimizes the number of segmented regions [53]. The marker controlled watershed algorithm along with the different features (colour, edge, orientation and texture) combinations was used for the extraction of tumor on brain MR images [54].
Clustering approach
In Data mining clustering is the technique of grouping homogeneous data into cluster based on some similarity criteria. The basic clustering segmentation algorithms in image processing are K-means clustering (hard clustering approach) and Fuzzy C means clustering (soft clustering approach). In Fuzzy C-Means (FCM) algorithm, the pixel/voxel can belong to more than one class and the fuzzy membership function value is the deciding authority to accommodate a pixel in the class. The membership function value lies in the range of 0 to 1 and if its value is one, the pixel at that location is very close to the centroid of the class and it can be accommodated in that class. The Euclidean distance is the commonly used distance metric to evaluate the creation of optimum number of clusters and its value should be minimum [55]. The conventional FCM algorithm is sensitive to noise and hence improvements in FCM were used in practice. Bias Field Corrected Fuzzy CMeans clustering (BFCM) algorithm corrects the intensity inhomogeneity in MR images produced by image acquisition system [56]. The improvements in FCM algorithm was done by modifying the distance measurements to allow the labeling of a pixel influenced by other pixels and for noise insensitivity [57,58]. Some of the improvements in conventional FCM algorithm are Generalized Spatial Fuzzy C-Means clustering (GSFCM), Mean shift based Fuzzy C-Means Clustering (MSFCM), Intuitionistic Fuzzy C-Means (IFCM), Type II Fuzzy C-Means (T2FCM) [59], Fuzzy Local Information CMeans clustering (FLICM) [60], Novel Fuzzy C-means clustering (NFCM), Improved Spatial Fuzzy C-means clustering (ISFCM) [61]. In [62], the authors brought to literature that, the variants of FCM clustering algorithm has been applied on the synthetic and real MR images. In the presence of noise, Fuzzy Local Information C-Means (FLICM) gives efficient result followed by Possibilistic Fuzzy C-Means (PFCM), Weighted Image Patch Based FCM (WIPFCM) and Multi-dimensional Fuzzy C-Means (MDFCM) algorithms. The Ant Bee Colony based FCM algorithm was used to determine the optimum cluster center. The proposed algorithm was tested on synthetic, medical and texture images, efficient result was produced when compared with GA, and PSO based FCM and EM algorithm [63]. [64] clearly put forth the Adaptively Regularized Kernel-Based Fuzzy C-means (ARKFCM) clustering for segmentation of magnetic resonance images of brain and it produces robust result in the presence of noise with low computational complexity. In [65], the local neighbourhood details and phase congruency features were used to define isotropic or anisotropic neighbourhood configuration of pixels, incorporated in the conventional FCM for accurate and noise robust image segmentation.
Markov random field models
Markov Random Field model have been widely used in image processing for segmentation and restoration of image, since it can preserve the edges by parameter estimation [66]. Markov Random Field (MRF) has prior information of index probability distribution of adjacent pixels. Ising Model and Potts Model are the standard MRF models, which solves many problems in image analysis [66]. The Hidden Markov Random Field (HMRF) model concept is derived from Hidden Markov Model (HMM). The HMRF model consist of two stochastic processes in which state sequence is not directly observable in the underneath stochastic process, but only through a sequence of observations [67]. In HMRF the segmented image can be considered as the realization of Markov Random Field ‘M’ defined on the same lattice ‘R’; taking values in the discrete space π={1, 2,…K}, where K represents the number of classes or homogeneous regions in the image. HMRF model is mathematically categorized as Hidden Random field, observable Random field and conditional independence. HMRF model is efficient in multi-dimensional (2D/3D) segmentation, where the Markov chain in Hidden Markov model is helpful for one-dimensional image segmentation as it is a first order neighborhood system [67]. The image segmentation in HMRF is modeled, as an optimization problem and the commonly used estimation technique are Maximum a Posteriori (MAP) or Maximum Likelihood (ML) principles [68]. The HMM model based on Expectation Maximization (EM) algorithm was applied to the MR images there by correcting the intensity inhomogeneity caused by the nonlinear response of the RF coil. The improved HMM-EM model produces robust segmentation result for MR images taking in to account of the partial volume effect [67]. The Region Based Hidden Markov model (RHMM) incorporates the anatomical information; the probability of voxels is assigned with respect to gray matter, white matter and cerebrospinal fluid in the case of MR images of brain [69]. In [70], the authors proclaimed Pickard random fields (PRFs) an unsupervised MRF model for the segmentation of breast mass. The PRF model was found to be efficient when compared with conventional MRF in terms of computational complexity.
Third Generation Segmentation Algorithms
Atlas based image segmentation
The atlas is an image comprising of anatomical details to incorporate prior information for segmentation and the results varies based on the atlas information [71]. The data sets of clinically ill subjects and some times normal subjects are used for the construction of atlas [71]. The atlas can be termed as the manually labeled image, which has close relation with the image to be segmented. The image registration plays a vital role and multiple atlases improve the segmentation accuracy [71]. During the image registration, the manually labeled atlas was transformed by mapping technique often termed as label propagation to accurately segment the target object [72]. The selection of reference images for atlas construction was done by either picking the sample closer to the mean or to determine true mean of the population [73]. A wide number of iterative algorithms are there based on several parameters to reduce the bias effect in the atlas construction. In the case of multiple atlas segmentation, database will be large and the selection of appropriate atlas for the query image is done [73]. The accuracy of registration is vital since error will occur, when there is a topological difference in the atlas and query image [72]. The labels of atlas images for target delineation was framed by spatially varying decision fusion weights derived from local assessment of the registration process [74]. The proposed average-shape atlas-based segmentation method yields better result than single atlas-based technique on cardiac and aortic segmentation in CT scan images [74]. It became factual from [75] that for the analysis of ventricular and atrial functions, cardiac chamber segmentation was done by fully automatic atlas based approach on 3D Computed Tomography Angiography (CTA) images. In the citation [76], the authors plotted that the statistical atlas was used for the initialization of robust shape based deformable model to segment 3D liver in MR images. Sema et al. [77] laid out an idea that the combination of manually delineated model along with simulated models determined from CT scans and bone-tissue images from dual energy X-ray machines were used for the automatic segmentation of rib bones in chest X-rays.
Artificial neural networks
Artificial Neural Network has already made a strike widely in the areas of Medical image processing. Artificial Neural Networks are mostly used for segmentation and classification by the adaptive learning approach. Artificial neural networks are represented by a set of nodes, often arranged in layers and a set of weighted directed links connecting them [78]. The nodes are the information processing units and the links acts as communicating media. There are a wide variety of neural networks depending on the nature of information processing carried out at individual nodes, the topology of the links, and for adaptation of link weights [79]. The two important aspects in neural network are training and learning. The neural network is trained with the features which may be statistical features such as mean, standard deviation, kurtosis, skewness or transform based features by applying wavelet transform or curvelet transform. The initial stage of neural network during training can be termed as guessing stage and as the training proceeds, stable state will be attained. More the training provided to the neural network, better will be the result with respect to query input. Learning is an adaptive process in which the weights associated with the interlinking neurons change in order to provide best response. Based on the learning process, the neural networks can be classified into supervised learning and unsupervised learning technique [79]. Neural Network algorithm has the major complication in the selection of the different architecture, network size, type, number of layers and topologies. Selection of these factors affects the performance of processing. The GMDH type Neural Network aid in automatic selection of architectural design decisions and thus solves the problem of prior knowledge about the system [80]. Neural network based on fuzzy logic are used in wide areas of medical image applications like tissue classification, anomalies detection in mammogram images, MR brain images etc. [81]. Fuzzy neural networks are broadly classified into three categories concurrent fuzzy neural system, cooperative fuzzy neural networks and hybrid techniques. Fuzzy neural networks are insensitive to noise and provide better segmentation result. The wavelet based neural network is widely used in medical image segmentation, compression, classification [81]. An interactive segmentation algorithm based on Support Vector Machine (SVM), Kth Nearest Neighbor (kNN) and decision tree was applied for the segmentation of brain tumor and kernel function based SVM produces better results [82]. It became factual from [83] that the single hidden layer feed forward neural network along with 3D fast marching algorithm was used for the segmentation of liver tumor from MR images of abdomen. The results were validated with ground truth image, the time complexity was greatly reduced and accurate results were obtained when compared with other semiautomatic segmentation approaches [83]. In [84], the KNN classifier produces good accuracy for Abdominal Aortic Calcification (AAC) detection in dual Energy X-Ray Absorptiometry images when compared with SVM technique. The authors of [85] were diligent in their studies that the Deep Convolution Neural Network (DCNN) based automatic brain tumor segmentation model incorporates local and global contextual features with two-phase training stages.
Graph cut approach
The graph cut segmentation algorithm is based on the graph theory in mathematics. A graph is represented by set of nodes or vertices and the connection between the neighboring vertices are depicted by edges. In the case of representation of image as a graph, nodes are used to characterize the pixels and the edges are the link that connects the pixel, whose weights corresponds to the cutting cost [86]. The S (source) and T (sink) are the special nodes that depict the object and background seed points [86]. The n links are the edges that connect between neighboring nodes and t links are the edges that connect between pixels of terminal nodes. The O and B represents the object and background seeds such that O ∩ B=Ø. The main objective of graph cut algorithm is to perform an optimal cut that separates the object and background [87]. The prior knowledge of the shape of the object (desired ROI) can be included in the graph cut algorithm to produce robust result [88,89]. The shape priors graph cut segmentation algorithm produce optimum results than conventional graph cut algorithm. The star shape prior graph cut model includes an objective function based on the balloon term so that larger object segmentation can be done [88]. The graph cut algorithm is also efficient for multi object segmentation in 3D images [89]. In [90] the kernel graph cut method was found to be good in the multi region segmentation of synthetic and MR images of brain. The RBF kernel based graph cut techniques comprises of graph cut optimization and finite intervals for updating the region parameters. In [91], the authors proposed a less computational complexity interactive graph cut model based on circular template for the segmentation of liver tumor on US images. The authors of [92], were studious in their views that the hierarchical graph segmentation based on perceptual Gestalt principles (e.g., similarity, closure, proximity, and continuity) was used for the extraction of blood vessels in retinal fundus images.
Hybrid approaches
In the recent years, the hybrid segmentation approach plays a vital role in many applications to extract the desired of interest. A hybrid technique comprising of conventional watershed algorithm and FCM along with fuzzy relations was developed to eliminate the over segmentation which is an inherent problem in watershed algorithm [93]. The optimization tool in graph cut algorithm and the iterative contour evolution of level set are utilized to formulate a hybrid algorithm (Graph cut based active contour) for the segmentation on 2D and higher order dimension images [94]. The watershed and threshold algorithm was combined for the efficient segmentation of tumors in the MR images of brain [95]. The region growing and level set algorithms along with artificial neural network were used for the segmentation and classification of breast tumors [96]. The local Chan-Vese model and finite Gaussian mixture model was combined such that it yields efficient results for low contrast and noisy images [97]. The Otsu thresholding technique was modified by flower pollination algorithm with modified randomized location and it produces better results than conventional thresholding technique [98]. The issues in conventional segmentation algorithms are solved by hybrid approaches by incorporating the desired features like spatial information, shape prior information etc. In [99], the authors were meticulous in giving an idea of neural network based watershed algorithm, the watershed with multilayer perceptron extracts the liver region in one slice and the boundary tracking algorithm was used to extract the liver region in other MRI slices. The features extracted from the image were used to set the parameters in the watershed algorithm; there by the over segmentation problem in conventional watershed algorithm was minimized. In [100], the authors contemplated on the localized active contour model along with multi thresholding based on krill herd bio inspired optimization algorithm which has been applied for the segmentation of nuclei in stained breast biopsy images from MITOS dataset. In [101], aforethought on a hybrid segmentation algorithm comprising of mean shift clustering and level set model was proposed for medical images with novel functions to control the parameters of the level set model based on the clustering result. Hayder et al. in [102] regarded that the HMRF model along with thresholding was used for the segmentation and quantification of brain tumor in MR images. Stavros et.al [103] reasoned the registration and segmentation on MR images of brain was done by a unified approach comprising of atlas and pair wise Markov Random Fields. Lu et al. in [104] put forth that the convolution neural network was used for the detection of liver from CT images and the result was refined by graph cut algorithm.
Results and Discussion
This section enumerates the experimental results of some of the typical segmentation algorithms on abdomen CT images. The simulation is attained with MATLAB 2010a on a desktop computer with 64-bit OS running on Intel Core i3-2120 CPU at 3.30 GHz with 4.00 GB RAM and integrated Graphics processor. The CT images are acquired from Optima CT machine with 0.6 mm slice thickness. The abdominal CT images of 4 data sets were used and the pathological information is depicted in Table 1. The ethical committee in Mar Ephraem Center for Medical Image processing and Metro Scans and Research Laboratory, Thiruvananthapuram approved the study of CT images of human subjects for research work.
| Dataset ID | x/y/z (mm) | Dimensions | Pathological information |
|---|---|---|---|
| 1 | 0.6 | 512 × 512 | A well-defined hypodense cystic area with imperceptible wall suggestive of benign cyst in right lobe of liver. |
| 2 | 0.6 | 512 × 512 | A moderate sized mass lesion measuring 5.8 × 3.2 cm is noted in segment-VII of right lobe with puddling of contrast suggestive of benign tumour-hemangioma. |
| 3 | 0.6 | 512 × 512 | Coronal images showing the enhancing normal kidneys in the nephrographic phase. |
| 4 | 0.6 | 512 × 512 | A moderately enhancing mass lesion measuring 3.4 × 2.8 cm is noted in the upper pole anterior cortex of the left kidney suggestive of malignant mass lesion-renal cell carcinoma. |
Table 1. Pathological information of datasets.
The inferences made from the research papers about the different generations of segmentation algorithms are as follows:
First generation algorithms: The thresholding algorithm is computationally inexpensive and fast, but the segmentation result is influenced by noise, partial volume effects and hence edge preservation is poor. The region based algorithms are effective for well-defined regions, but the selection of seed point is crucial and it involves little time complexity. The edge based algorithms are easy to implement and fast, but not appropriate for extracting the desired ROI characteristics. The first generation algorithms play a vital role in the hybrid segmentation approach to yield a refined result. The thresholding and region growing algorithms are modified by incorporating the spatial local information, connectivity and by employing optimization techniques.
Second generation algorithms: The deformable models are widely used in case of complex geometry or large shape variation and are appropriate for boundary detection. The selection of parameters is crucial in deformable models and they are time consuming, requires manual intervention. The watershed algorithm is efficient when a local minima exactly corresponds to ROI and for uniform objects in the image; however there is a chance of over segmentation and missing of boundaries in low contrast images. The issues in conventional watershed algorithm were resolved by appropriate preprocessing and post processing technique. The HMM gives more emphasis on spatial information; however it is much applicable for MR images of brain and aerial images. The computational complexity of HMM is high and parameter selection for controlling the strength of spatial relations is difficult. The clustering algorithms are easy to implement since they does not require training phase, however to make it insensitive to noise, prior information about local statistics of pixels have to be incorporated.
Third generation algorithms: The classifiers are having good parallelism; however, the spatial modelling is poor and hence produces issues in the segmentation of MR images with intensity inhomogeneity. The classifiers based on supervised approach need good number of samples for training to yield accurate segmentation result. The graph cut is a special case of MRF and unlike active contour models, the computational complexity is reduced. The automatic seed point selection in graph cut needs the shape prior information and a hybrid technique comprising of neural network with graph cut will yield good results. The atlas-based approach is optimum for segmentation of structures that are consistent in the population of study when a standard atlas or template is available, still the anatomical variability and ROI of complex composition are the issues. The third generation algorithms are made insensitive to noise by incorporating neighborhood and geometric information, though these algorithms requires manual interaction and are time consuming.
The hybrid segmentation algorithms combine the algorithm in each generation to yield good results. The medical image characteristics and ROI topology have to be given importance while formulating a hybrid segmentation model. The segmentation results of some of the typical algorithms in each generation are depicted below. The pathological information of CT images is shown in Table 1.
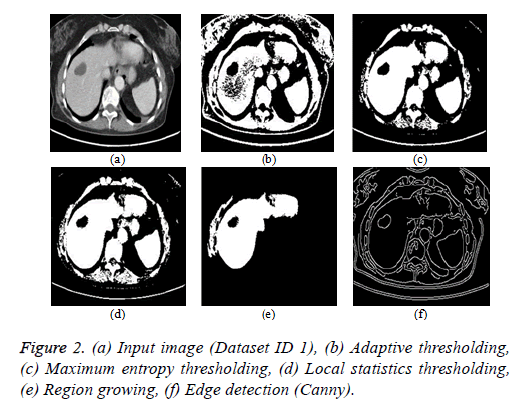
Thresholding algorithm: The thresholding is a basic segmentation algorithm and the result of binary Thresholding is shown in Figure 2b. The threshold value of T=0.65 is chosen based on the histogram analysis. The maximum entropy Thresholding algorithm result is shown in Figure 2c with a maximum threshold value (hmax) of 5.2735. The result of thresholding based on local statistics is depicted in Figure 2d. The parameters of the algorithm are kernel size (3 × 3), nonnegative scalar integer values (a=2 and b=1.5).
Region growing algorithm: The result of the conventional region-growing algorithm with manual seed point selection corresponding to Figure 2a is depicted in Figure 2e. The seed point value of (x, y) is (135, 105) with a threshold of T=0.2 is chosen and the region growing process terminates when the intensity difference between the region mean and new pixel becomes greater than the threshold.
Edge detection algorithm: The different types of edge detection algorithms are Sobel, Canny, Prewitt and Roberts in which Canny produces better results compared with other edge detection operators and the result is depicted in Figure 2f.
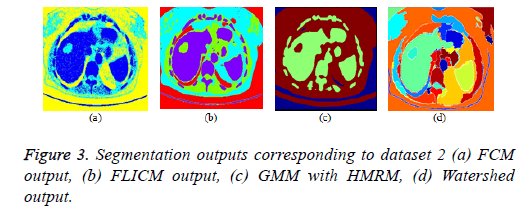
FCM clustering algorithm and its variants: The parameters of the FCM algorithm are cluster fuzziness (f), number of clusters (c) and stopping criterion (ε). The cluster fuzziness (f=2), number of clusters (c=3) and stopping criterion (ε=0.001) are employed for the FCM algorithm. The cost function minimization takes place by comparing the changes in the membership function or the cluster center at two consecutive iterations. The cost function minimization takes place when the change in consecutive values of fuzzy membership function is less than the stopping criterion (ε=0.001). The FCM output is depicted in Figure 3a. The conventional fuzzy clustering algorithm is sensitive to noise; hence, a suitable preprocessing technique is needed prior to clustering. In FLICM algorithm, a new fuzzy local neighborhood factor (G) was used that explores the spatial and gray level relationship so that the clustering result is insensitive to noise, thereby it generates better result than conventional FCM algorithm. The FLICM algorithm eliminates the crucial parameter selection (α or λ) in the objective function of modified FCM clustering techniques [57,59,105,106] that makes it robust to noise. The parameters of the FLICM algorithm are Number of clusters (C=4), Fuzzy membership weight value (m=2), Size of the local kernel (w=3), Maximum number of iterations (i=500), Stopping criterion threshold value (thresh=0.001). The FLICM output is depicted in Figure 3b.
GMM based HMRF algorithm: In this paper, Gaussian based hidden Markov Random Field (HMRF) and its Expectation Maximization (EM) algorithm is used and the clustering result is better than K means Clustering algorithm. The parameters of the HMRF-EM framework with GMM model [107] are number of regions (k=3), number of GMM components (g=3), EM iterations (e=10), Maximum a posteriori probability (MAP) iterations (m=10) and the result is depicted in Figure 3c.
Marker controlled watershed algorithm: The over segmentation problem in conventional water segment algorithm was eliminated by marker controlled watershed algorithm (foreground and background marker). The marker controlled watershed algorithm along with morphological operations is employed in this paper and the result is depicted in Figure 3d.
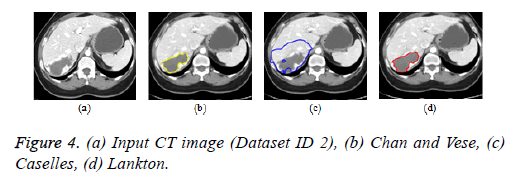
Active contour models: The Creaseg software comprising of different level set algorithms was used for the boundary detection of liver in abdomen CT images [108]. Three level set algorithms are taken into account comprising of Chan and Vese, Caselles and Lankton active contour model. The Caselles active contour model uses gradient of the image to determine the energy function and the curve is evolved into the regions of high gradient [109]. The Chan and Vese model is a region based model in which the image is subdivided into two homogeneous regions based on the energy constraints and the curve evolution is based on the narrow band of level set [110]. The Lankton algorithm [111] evolve the initial contour based on the local neighborhood statistics and can segment the image into two homogeneous regions; the results are depicted in Figures 4b-4d. The Lankton algorithm produces superior results when compared with Chan and Vese and Caselles model.
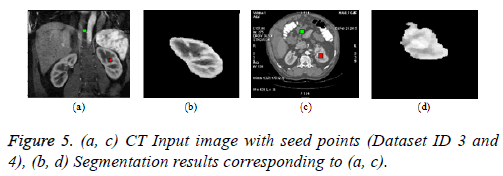
Graph cut algorithm: The Boykov-Kolmogorov graph cut algorithm is used in this paper for the segmentation of kidney from abdomen CT images [8]. The BK algorithm comprises of three stages growth, augmentation and adoption stage. The seed point selection is crucial and it should be done properly for accurate segmentation. The result can be enhanced by incorporating neural network for automatic seed point selection by proper training. The Figure 5b below depicts the result of graph cut algorithm for the segmentation of kidney from normal coronal slice and Figure 5d depicts the segmentation of kidney with malignant tumor from axial slice.
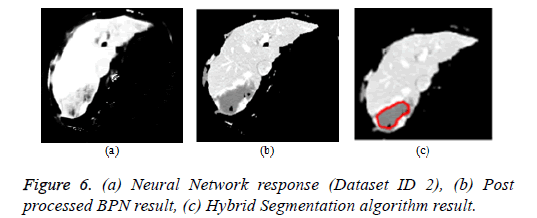
Artificial neural network: The back propagation neural network is widely used in pattern recognition and classification. For the segmentation of liver from abdomen CT images, BPN is used in this paper [112]. The feature extraction comprises of local features (mean, variance, local minimum, local maximum) and texture features (contrast, correlation, energy and homogeneity). The BPN is trained with the features and the hidden layer comprises of 20 neurons. The third layer is the output layer that comprises of one neuron depicting ‘1’ for liver region pixels and ‘0’ for non-liver region pixels. The neural network response was post processed by morphological operations to produce the refined output. The Figure 6a depicts the neural network response and Figure 6b represents the neural network response after post processing by morphological operations.
Hybrid segmentation algorithm: The hybrid segmentation algorithms are now playing a major role in yielding better results. In this paper, the hybrid segmentation algorithm comprising of neural network and level set model is proposed. The BPN algorithm segments the liver from input image and the tumor region was outlined by localized region based active contour model [112]. The CREASEG software was used where many level set models are there, out of which the Lankton algorithm produces superior results and the hybrid algorithm result is depicted in Figure 6c.
The evaluation of segmentation result is vital and it is done by supervised or unsupervised techniques [113]. In supervised technique, the evaluation is done by comparing the resultant segmented image against a manually segmented reference image which is often called as gold standard or ground-truth image. The measure of resemblance between the human segmented image and machine segmented image determines the quality of segmented image [114]. In the case of unsupervised evaluation methods, no reference or ground truth image will be there and the evaluation is done by extracting the characteristics of the segmented image and comparing with the pre-set desired characteristics [115]. The creation of ground truth or reference image is not possible in all real time applications and it is time consuming, difficult. There are number of measures in unsupervised segmentation technique like entropy, region shape, intra region uniformity and inter region contrast which doesn’t need a reference image [116].
Conclusion
The different types of segmentation algorithms are discussed and the results of some typical algorithms are presented in this paper. It is factual that there is no universal algorithm for medical image segmentation, wherein, the choice depends upon the image modality, characteristics of region of interest and application. This review will be a vital tool for the researchers to choose an appropriate algorithm for their application. There is neither a single segmentation model for all medical image modalities nor all methods are efficient for a specific medical image modality. In image processing and computer vision, segmentation is still a challenging problem in many real time applications and hence more innovative work is required. This work will be a guideline for the researchers to develop new segmentation models.
Acknowledgement
The authors would like to acknowledge the support provided by the DST under IDP scheme (No: IDP/MED/03/2015). Also the authors are thankful to Metro Scans and Research Laboratory, Trivandrum for supporting the research work.
References
- Aldasoro CCR, Abhir B. Volumetric texture segmentation by discriminant feature selection and multiresolution classification. IEEE Trans Medical Imaging 2007; 26: 1-14.
- Olivier J, Cedric M, Jean JR, Romuald B, Hubert C. A supervised texture-based active contour model with linear programming. Proc 15th IEEE International Conference Image Processing 2008; 1104-1107.
- Schaap M, Theo W, Lisan N, Coert M, Ermanno C, Marleen de B, Wiro N. Robust shape regression for supervised vessel segmentation and its application to coronary segmentation in CTA. IEEE Trans Medical Imaging 2011; 30: 1974-1986.
- Lee NR, Theodore S, Andrew FL. Interactive segmentation for geographic atrophy in retinal fundus images. Conference Record/Asilomar Conference Signals, Systems and Computers. Asilomar Conference Signals Systems Computers 2008; 2008: 655.
- Grau V, Mewes AUJ, Alcaniz M, Ron K, Simon KW. Improved watershed transform for medical image segmentation using prior information. IEEE Transactions Medical Imaging 2004; 23: 447-458.
- Mondal T, Ashish J, Sardana HK. Automatic craniofacial structure detection on cephalometric images. IEEE Trans Image Proc 2011; 20: 2606-2614.
- Kubota T, Anna KJ, Maneesh D, Marcos S, Arun K. Segmentation of pulmonary nodules of various densities with morphological approaches and convexity models. Med Image Anal 2011; 15: 133-154.
- Boykov YY, Jolly MP. Interactive graph cuts for optimal boundary & region segmentation of objects in ND images. Eighth IEEE International Conference Computer Vision 2001; 1: 105-112.
- Russ JC, Woods RP. The image processing handbook (3rd edn.) CRC Press Addison-Wesley Publishing Company USA 1993.
- Ganesh JS, Srinivasa Rao V, Srinivas K. Enhanced noise type recognition using statistical measures. IOSR J Comp Eng 2012; 2: 19-23.
- Yousuf MA, Nobi MN. A new method to remove noise in magnetic resonance and ultrasound images. J Sci Res 2010; 3: 81.
- Buades A, Bartomeu C, Morel JM. A non-local algorithm for image denoising. Proc IEEE Computer Society Conference Computer Vision Pattern Recognition 2005; 2: 60-65.
- Hennenfent G, Felix JH. Seismic denoising with nonuniformly sampled curvelets. Computing Science Engineering 2006; 8: 16-25.
- Dawant BM, Alex PZ, Richard AM. Correction of intensity variations in MR images for computer-aided tissue classification. IEEE Trans Medical Imaging 1993; 12: 770-781.
- Pina XP, Bach CM, Kunz N, Just N, Gruetter R, Ph Thiran J. Fast bias field correction for 9.4 tesla magnetic resonance imaging. Proc 16th European Signal Processing Conference 2008; 2008; 1-5.
- Karch P, Zolotova I. An experimental comparison of modern methods of segmentation. Proceedings IEEE 8th International on Applied Machine Intelligence Informatics 2010; 2010; 247-252.
- Wu J, Skip P, Michael DN, Markad VK. Texture feature based automated seeded region growing in abdominal MRI segmentation. Proc IEEE International Conference Bio Medical Engineering Informatics 2008; 2: 263-267.
- Thakur A, Radhey SA. A local statistics based region growing segmentation method for ultrasound medical images. World Academy Science, Engineering and Technology. Int J Med Health Biomed, Bioeng Pharm Eng 2007; 1: 564-569.
- Nadeem M, Asadullah S, Ahmad W, Adamu A, Ahafia K, Syed BZ. Image segmentation methods and edge detection: an application to knee joint articular cartilage edge detection. J Theor Appl Inform Technol 2015; 71.
- Lopez-Molina C, De Baets B, Bustince H, Sanz J, Barreneche E. Multiscale edge detection based on Gaussian smoothing and edge tracking. Knowledge Based Systems 2013; 44: 101-111.
- Mesanovic N, Mislav G, Haris H, Matija M, Emir S, Muamer S. Automatic CT image segmentation of the lungs with region growing algorithm. Proc 18th Int Conference Systems Signals Image Processing 2011; 1: 395-400.
- Grenier T, Chantal RM, Costes N, Janier M, Gerard G. 3D robust adaptive region growing for segmenting [18F] fluoride ion PET images. Proc IEEE Nuclear Science Symposium Conference Record 2006; 5: 2644-2648.
- Lehmann TM, Daniel B, Christian T, Thomas S. Segmentation of medical images combining local, regional, global, and hierarchical distances into a bottom-up region merging scheme. Medical Imaging International Society for Optics and Photonics 2005; 546-555.
- Chuin MW, Shao WS, Pei CK, Geng CL, Da PY. A study on the application of fuzzy information seeded region growing in brain MRI tissue segmentation. Math Problems Eng 2014; 2014: 1-11.
- Samy S, Ayoub AH. Entropic image segmentation: a fuzzy approach based on Tsallis entropy. Int J Comp Vis Sig Proc 2015; 5: 1-7.
- Sheema SK, Gule S, Imran K, Abdus S. Maximum entropy based image segmentation of human skin lesion. World Academy of Science Engineering and Technology. Int J Comp Electr Autom Control Inform Eng 2015; 9.
- Asma K, Asma B, Antoine M, Mohamed HB. A coronary artery segmentation method based on multiscale analysis and region growing. Comp Med Imag Graphics 2016; 48: 49-61.
- Kass M, Andrew W, Demetri T. Snakes: active contour models. Int J Comp Vis 1998; 1: 321-331.
- Yeo SY, Xianghua X, Igor S, Perumal N. Geometrically induced force interaction for three-dimensional deformable models. IEEE Trans Image Proc 2011; 20: 1373-1387.
- Malladi R, James AS, Baba CV. Shape modeling with front propagation: A level set approach. IEEE Trans Patt Anal Mac Intel 1995; 17: 158-175.
- Xu C, Prince JL. Snakes, shapes, and gradient vector flow. IEEE Trans Image Process 1998; 7: 359-369.
- Laurent DC. On active contour models and balloons. CVGIP Image Understanding 1991; 53: 211-218.
- Tsai A, Anthony Y, William W, Clare T, Dewey T, Ayres F, Eric GW, Alan W. Model-based curve evolution technique for image segmentation. Proc IEEE Comp Society Conference Comp Vision Pattern Recognition 2001; 1: 1-463.
- He L, Zhigang P, Bryan E, Xun W, Chia YH, Kenneth LWand William GW. A comparative study of deformable contour methods on medical image segmentation. Imag Vis Comp 2008; 26: 141-163.
- Lankton S, Allen T. Localizing region-based active contours. IEEE Trans Image Proc 2008; 17: 2029-2039.
- Chan TF, Luminita AV. Active contour and segmentation models using geometric PDEs for medical imaging. Geometric Methods Bio-Medical Image Processing Mathematics Visualization Springer Berlin Heidelberg 2002; 63-75.
- Tsai A, Anthony Y, William W, Clare T, Dewey T, Ayres F, Eric GW, Alan