ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 17
A reverse genetic approach to study the pathogenicity of coxsackievirus A6
In recent years, CVA6 has become a new pathogen of Hand, Foot, and Mouth Disease (HFMD), associated with crusted lesions, a rash affecting the arms, trunk, and legs, nail loss. In order to dig out the unclear pathogenesis of CVA6-associated HFMD, we established a reverse genetic system for CVA6 and determined the virulence and the proliferation of the rescued virus. In this study, the CVA6 viral cDNA was divided into two segments, which were ligated through the BclI restriction site. The constructed plasmid was designated as pWSK29-CVA6-89R identified by enzyme digestion and the whole cDNA sequencing. The rescued virus CVA6-89R was infected in ICR mice, showing high pathogenicity. This study established a reverse genetic system for CVA6.
Keywords
Reverse genetic system, Coxsackievirus A6, Hand, Foot, Mouth disease
Introduction
Hand, Foot, and Mouth Disease (HFMD) is a common disease worldwide, mainly occurring in children under 3 y old [1]. The disease is caused by a variety of intestinal viruses and characterized by fever and herpes blisters on the hands, feet, mouth, and other body parts [2]. A few children suffer serious complications, including encephalitis, acute flaccid paralysis, myocarditis, and cerebral edema. HFMD progresses rapidly and sometimes can cause death in severe cases [3]. In recent years, there have been several outbreaks or epidemics of HFMD in many countries, especially in the Asia-Pacific region, and it has attracted increasing attention throughout the world [4-7]. The causative agents of the disease include a variety of enteric viruses, but the main pathogens are Enterovirus 71 (EV71) and Coxsackievirus A16 (CVA16) [8-10]. Moreover, cases of CVA6-induced HFMD are increasing year by year [11-13].
In the summer of 2013, HFMD broke out in Changchun, China. In total, 253 samples were collected and tested, and 158 (62.45%) were infected with CVA6 [14]. CVA6 had replaced EV71 and CVA16 as the main etiological agent of HFMD in Changchun. Nevertheless, CVA6-induced HFMD was not detected in Changchun (China) in 2012 [15]. Instead of EV71, CVA6 has become the major pathogen of HFMD in Guangdong, and other places since 2013 [16-19].
CVA6, a single-stranded enteric RNA virus with spherical and icosahedral symmetry particles, is mainly spread by the fecaloral route and is one of the pathogens of HFMD [20,21]. Since 2013, CVA6-caused HFMD has shown an increasing trend [22]. In east China, densely populated areas of the country, the disease seriously threatens the life and health of children. Thus, the prevention and treatment of coxsackievirus is quite a challenging job.
To better prevent the disease and control the virus responsible, the transmission mechanism and pathogenesis of the etiological agent must be investigated. Reverse genetics has been widely used to study the pathogenicity, transmission capacity, and mutation mechanisms of influenza virus. In this study, we first established a reverse genetic system for CVA6, at the level of its genes and proteins. Animal experiments were also used to determine the virulence of the rescued virus. The reverse genetic system developed for CVA6 in this study laid the foundation for further research on the gene functions, pathogenesis, and transmission mechanism of CVA6.
Materials and Methods
Viruses, plasmids, and cells
The reagents used were: Lipofectamine™ LTX with Plus Reagent (Invitrogen), human malignant embryonic Rhabdomyoma (RD) cells (Shanghai Cell Institute of Chinese Academy of Sciences, China), fluorescent CVA6 probe (provided by the Zhejiang Provincial Center for Disease Control and Prevention), T vector (TaKaRa Biotechnology (Dalian) Co., Ltd, China), pWSK29 (ampicillin-resistance lowcopy plasmid; a gift from researcher Mao Pan-yong, 302 Military Hospital of China), double-antibody penicillin– streptomycin, and trypsin digestion reagent (Beijing Suolaibao Biotechnology, China), mouse anti-CVA6 antibody (Shanghai Enzyme-linked Biotechnology Co., Ltd, China), and horseradish-peroxidase-labeled goat anti-mouse IgG antibody (Beyotime Biotechnology, China). Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum, Phosphate- Buffered Saline (PBS) was purchased from Thermo Fisher (American).
Virus origin and identification
Throat swab samples were collected from a child with HFMD, who was identified in the Jiaxing Center for Disease Control and Prevention of China. CVA6 Serotype of the specimens was identified by RT-PCR.
Construction of a plasmid containing all the viral genes
After infection with 10 CVA6 strains, the death rate results suggested that the toxicities of ten virus strains were remarkably different. The toxicity of CVA-89 was strongest. The CVA6-89 VP1 region of the isolated strain was sequenced, and a GenBank sequence alignment confirmed to be the CVA6-TW subtype (type CVA6A). The LaserGene software was used to design two pairs of whole-genome-amplifying primers and four pairs of identified primers, based on the conserved CVA6-TW sequence in GenBank. The isolated virus was a single-stranded RNA virus, with a full length of 7,460 bp. The 5’ end of primer 1 F introduced an EcoRI restriction enzyme site (GAATTC) and the T7 RNA polymerase promoter sequence. The 3’ end of primer 2 R introduced a BamHI restriction enzyme site (GGATCC). Using the T vector as the cloning vector for the rescue system, the viral cDNA was divided into two segments and each was cloned into the T vector. The primer sequences were showed in Table 1 and the primers were synthesized by Sangon Biotech (Shanghai) Co., Ltd, China. After the two gene segments were cloned, the entire cDNA was ligated into plasmid pWSK29 through the BclI restriction site shared by the two genome segments. Thus, plasmid pWSK29-CVA6-89R containing the full-length cDNA of the CVA6 strain was constructed. The plasmid pWSK29- CVA6-89R was confirmed with enzyme digesting identification by EcoRI and BamHI using an unrelated plasmid as the negative control. Plasmid pWSK29-CVA6-89R was sequenced by Sangon Biotech (Shanghai) Co., Ltd, China.
| Primer name | Sequence |
|---|---|
| 1 F | CGGGACTAATACGACTCACTATAGTTAAAACAGCCTGTGGGTTG |
| 1 R | GTCATAATCA CTTCTGATCA CTATGACTAA |
| 2 F | TTAGTCATAG TGATCAGAAG TGATTATGAC |
| 2 R | AGGATCCTTTTTTTTTTTTTTTTTTTTTTTTTTTCTGCTATTCTGGTTATA |
Table 1. Primers used for RT-PCR to synthesize two overlapping cDNA clones for CVA6 virus.
Virus rescue
RD cells were incubated in a six-well plate for 24 h, washed twice with PBS, and incubated with 1 ml of Opti-MEM at 37°C under 5% CO2 for 1-2 h before transfection. The RD cells were transfected with the constructed pWSK29- CVA6-89R plasmid. The pWSK29-CVA6- 89R plasmid (2 μg) was directly mixed with 500 μl of Opti-MEM. The mixture was incubated with 4 μl of Plus Reagent at room temperature without stirring for 10 min. Lipofectamine™ LTX (10 μl) was then added and allowed to mix with the cells at room temperature without stirring for 30 min. The mixture was incubated in a six-well plate at 37°C under 5% CO2 for 6 h. After the supernatants were discarded, the samples were incubated with 2.5 ml of Minimal Essential Medium (MEM) containing 10% fetal bovine serum for 72 h, and then at 37°C under 5% CO2 until pathological changes were observed in the cells. We set up four groups simultaneously: cells transfected with the pWSK29 plasmid, normal untransfected cells (negative control), pWSK29-CVA6-89R plasmid, CVA6-89- infected cells (positive control).
Western blotting assay of viral protein expression
RD cells showing a dramatic CPE were frozen and thawed three times. The pWSK29-CVA6-89R-infected-cell supernatants were added to 5X SDS loading buffer. After they were boiled for 5 min, the samples were rapidly cooled on ice and resolved electrophoretically at an initial constant voltage of 100 V. The voltage was adjusted to 120 V after the sample entered the stacking gel. After electrophoresis, the proteins on the polyacrylamide gel were transferred to polyvinylidene difluoride membrane at 250 mA and 4°C for 120 min. A mouse anti-CVA6 antibody was used as primary antibody and a horseradish-peroxidase-labeled goat anti-mouse IgG antibody was used as the secondary antibody. Cells were transfected with the pWSK29 plasmid and normal untransfected cells as negative control, and cells were infected with CVA6-89 as positive control.
Determination of the median tissue culture infective dose (TCID50)
The virus solution was diluted 10-fold, from 10-1 to 10-10. The diluted virus was seeded on a 96-well plate. Each dilution of the virus was seeded in a row (a total of eight wells), with 100 μl in each well. The cell suspension (100 μl) was added to each well at a final concentration of 2 × 105 cells/ml. Normal cells were added to two rows and used as the controls. The results were observed and recorded for 7 d. The results were calculated with Karber’s method, as logTCID50=L-(s-0.5), where L is the log value of the highest dilution of the virus; D is the difference between the logarithm of the virus dilution; and S is the sum of the positive ratio.
Construction of the viral growth curve
When the RD cells grew well and formed a monolayer in a 3 ml culture flask, the culture medium was discarded and the virus solution was added (3 ml/culture flask, total of 14 culture flasks), seven culture flasks were used as the controls. All the samples were incubated in a CO2 incubator at 37°C. Two flasks of virus-infected cells and one flask of control cells were harvested every 24 h and frozen at -80°C for 7 consecutive days. The virus was purified with the QIAamp Viral RNA Mini Kit. The viral load was determined with real-time PCR.
Mice infection
In vivo ICR mice (10 females, 4-5 w; 10 males, 4-5 w) were purchased from Zhejiang Academy of Medical Sciences, China (animal license number: SCXK (Hu) 2013-0016). The first generation of suckling mice was used for this study. Animal experiments were approved by the Ethics Committee of Jiaxing University, China. 1 d old ICR suckling mice were intracranially injected with CVA6 strain at the dose of 103TCID50. The rescued virus and parental virus at the dose of 102TCID50 were intraperitoneally injected into 1 d old ICR suckling mice. PBS was injected into the controls. Preinfection was considered as d 0. After injection, we observed the changes in food intake, activity, and mental state daily for 1 consecutive week. Mouse weight was weighed at 0, 2, 4, 6, 8, and 10 d after inoculation. Mice were anesthetized using ether inhalation. Two mice were sacrificed by euthanasia, and then the lung, brain and spleen were collected.
Virus loads in mouse tissues
At 3, 5, and 7 d after virus inoculation, lung, brain and muscle were collected from the CVA6-89, CVA6-89R, and control groups. After 14 d of observation, lung, brain, intestine, hind limb muscles were obtained to extract total RNA with RNeasy Mini Kit (Qiagen). RT-PCR was used to amplify CVA6 virus VP1 region gene. Primer sequences are as follows: CVA6 (sense) 5’- TCGAAATGGGGTTAATGAGGCG3’, (anti-sense) CRCCTTCATAATCMGTGGTGG. RT-PCR results were measured with agarose gel electrophoresis.
Statistical analysis
The data were analysed with the SPSS 18.0 software. Intergroup data were compared with a t test, and differences were deemed significant at α=0.05.
Results
Construction of plasmids containing the full length CVA6 viral genes
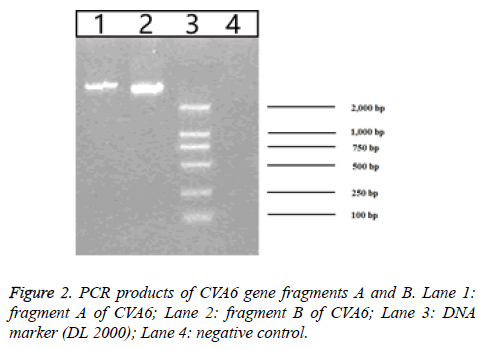
157 throat swab samples of HFMD patients were detected by using fluorescent RT-PCR. The result showed that the positive rate of EV71 was 29.29% (46/157), the positive rate of CVA16 was 28.66% (45/157) and the positive rate of CVA6 was 21.02% (33/157). The construction strategy of the plasmid pWSK29-CVA6-89R is listed in Figure 1. Based on the CVA6-89 gene sequence was over 7000 bp, RT-PCR was used to amplify two segments of the CVA6 genome. They were separately cloned into the T vector to generate stable clones. Gene segment A was digested with EcoRI and BclI and segment B of the cDNA was digested with BclI and BamHI. The two segments of the genome were successfully inserted into plasmid pWSK29, and the resulting plasmid was designated pWSK29-CVA6-89R. After enzyme digesting identification, 1% agarose gel electrophoresis revealed that the enzyme digesting fragments were 4,000 bp and 3,460 bp as expected (Figure 2), confirming that the full-length cDNA of CVA6-89 was inserted into vector pWSK29. These results demonstrated that the reverse genetic system plasmid pWSK29-CVA6-89R was successfully constructed.
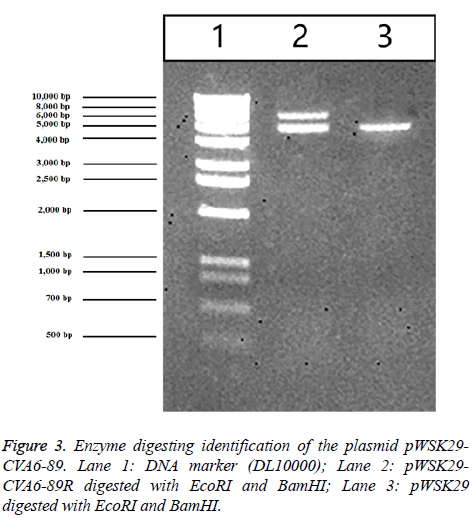
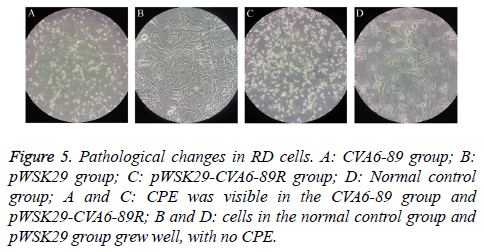
RD cells were separately transfected with the constructed pWSK29-CVA6-89R and the pWSK29 plasmid with untransfected cells (negative control) and CVA6-89-infected cells (positive control). After 96 h, pathological changes were visible in the cells of the pWSK29-CVA6-89R group and CVA6-89-infected group, and partial cell loss and cell shrinkage were seen under a microscope. The cell death rate was more than 90% in the rescued virus pWSK29-CVA6-89R group and the CVA6-89 virus group. However, the cell morphology was normal in the untransfected group. In the culture plate transfected with pWSK29 plasmid group and untransfected cells, the cells were round, smooth, showed a clear boundary, and grew well. In the culture plate transfected with pWSK29-CVA6-89R group and CVA6-89-infected group, the well grown cells had shrunk, and displayed strong refraction, with increased numbers of particles in their cytoplasm (Figure 3). Some cells were botryoidal and detached. These phenomena were typical of the virus-induced CPE. These cell infection results suggested that the constructed reverse genetic system of CVA6-89 can induce pathological changes in RD cells as the parental viruses.
Viral protein expression detected with Western blotting
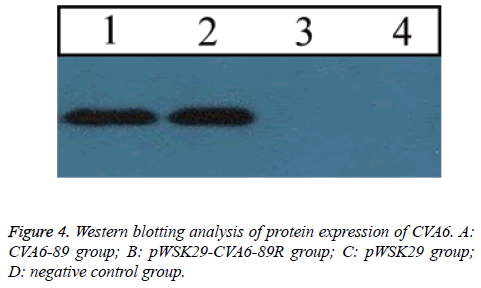
In RD cells with CPE+++, a Western blotting assay revealed a distinct protein band produced by the pWSK29-CVA6-89R group (Figure 4). A protein band was also observed in the same position in the CVA6-89 group. However, no protein band was seen in the normal control group and pwsk29 group. These findings suggested that the rescued virus properly expressed the viral proteins, which were immunogenic.
Viral growth curves
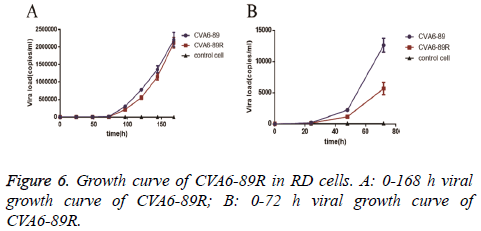
The viral titer (TCID50) was measured three times and was 10-3.875/0.1 ml for the rescued virus group and 10-3.375/0.1 ml for the CVA6-89 virus group. To compare the proliferation of the rescued and CVA6-89 viruses in RD cells, the same batch of RD cells was inoculated with the same titer (10 TCID50) of the viruses. After 24, 48, 72, 96, 120, 144, and 168 h, the viral loads were determined and growth curves drawn (Figure 5). The experimental results demonstrated that the viral load was noticeably increased at 48 h, and then increased exponentially. In the first 144 h, the numbers of rescued virus were lower than the numbers of CVA6-89 virus. However, after 144 h, there was no difference in the loads of the rescued virus and the CVA6-89 virus. The viral loads could not be monitored beyond 168 h because of cell loss. The growth curves showed identical trends in the proliferation of the rescued virus and the CVA6-89 virus (Figure 6).
Pathological changes and tissue viral loads in mice
At 3 d after injection with rescued virus and parental virus, suckling mice presented decreased activity, listlessness, hind limb paresis, even paralysis. Above symptoms lasted for 7 d, and then disappeared. Suckling mice in the control groups did not present any symptoms. In the CVA6-89R group and CVA6-89 virus group, virus was detected in the mouse brain and lung at 6 d after infection, detectable in the mouse lung, brain and spleen all three tissues at 8 d. However, virus was not found in the PBS group for all the time. RT-PCR detection results in ICR mice infected with the virus were showed in Table 2.
| Group | Control | CVA6-89 | CVA6-89R |
|---|---|---|---|
| Tissue where RNA is first detected | Not detected | 3 | 3 |
| Time of first detection of RNA (d) | Not detected | Brain, lung | Brain, lung |
| Time when RNA can be detected in all three tissues | Not detected | 8 | 8 |
Table 2. RT-PCR results in ICR mice after infection with CVA6 virus.
Discussion
In May 2008, CVA6-induced HFMD was first reported in Finland [23]. Since then, the incidence of CVA6-caused disease has increased each year, and CVA6 has become a main etiological pathogen instead of EV71 and CVA16 in some years and some areas. Unlike EV71 or CVA16, CVA6 does not contribute to severe nervous system symptoms, but the rash and fever symptoms are more serious in CVA6-infected patients [24,25]. From November 2011 to February 2012, California CDC reported that 76% of 63 CVA6-infected patients suffered fever, 67% were affected by rash on the hands or feet, and 41% by rash on the face [26]. Outbreaks of HFMD are gradually increasing worldwide, warning us to study the pathogenicity of CVA6 urgently, as well as to get the rescued CVA virus.
Based on the reverse genetics of RNA viruses, we tried to make some efforts to curb the prevalence of HFMD caused by CVA6. In 1981, the Racaniello et al. successfully constructed an infectious clone of poliovirus, which was the first successfully rescued RNA virus of animals [27]. A reverse genetic system can be used to investigate the pathogenicity and transmission capacity of a virus, as well as the mechanism of its variability, whose technique has become an important method for studying HFMD [28]. CVA6 is a single-stranded RNA virus that can rescue infectious CVA6 in vitro [29] and can provide a convenient way of analyzing and comparing the virulence and immunogenicity of viruses, as well as clarifying their infection mechanism and pathogenesis. This study generated CVA6-89 from the CVA6-89 virus, which was identified at the Jiaxing Center for Disease Control and Prevention of China with a fluorescent probe. After PCR amplification of the VP1 region, CVA6-TW was confirmed with a DNA sequence analysis. Using the long fragment and a high-fidelity Taq DNA polymerase, we amplified the whole cDNA of CVA6-89 as A and B segments and constructed a cloning vector. Increasing the polyA length in the 3’ untranslated region of the intestinal virus during the primer enhances the synthesis of the negative strand of the viral RNA. Therefore, the addition of 20 T nucleotides was expected to increase the synthesis of viral RNA. Simultaneously, the full use of BclI restriction sites of 3,971 sites in the CVA6 sequence connected the amplified A and B segments. Thus, an infectious clone containing the whole cDNA of the CVA6 virus was generated.
To investigate the virulence of the rescued virus, RD cells were transfected with the constructed infectious clone of pWSK29- CVA6-89R. The cells transfected with the rescued virus or the CVA6-89 virus showed marked CPEs 72 h after transfection. Identification of the genes and proteins of the rescued viral strains suggested that the rescued virus and CVA6-89 virus have similar biological properties. The constructed rescued virus infected RD cells, maintained a constant copy number in the cells, and induced pathological changes in the cells. Measurement of the viral titers revealed that the titer was 10-3.785/0.1 ml for the rescued virus and 10-3.375/0.1 ml for the CVA6-89 virus. The growth curves showed that virus numbers had no difference in the loads of the rescued virus and the CVA6-89 virus at 168 h.
In accordance with quantitative PCR, cytopathic effect test and challenge test, among 157 virus strains, CVA6-89 strain with strongest virulence was selected as a parental virus for establishing dCVA6 reverse genetics system. Identification of vectors and determination of viral load in different tissues of mice confirmed that rescued virus and parental virus had the same infectivity.
In this study, the parental virus was derived from a 5 y old male child with nail shedding, the infectious clone was constructed, after sequencing, 10 amino acid mutations were detected in the infectious clone. Compared with the CA6 strain TW-2007-00141, three amino acids were mutated in 5’ UTR region of the parental virus, two amino acid mutations were found in 3D protein structure. 5’ UTR mutations were associated with viral replication and virulence, while the amino acid mutations in 3D protein might affect the structure of the CVA6 RNA polymerase, and then influence RNA polymerase activity and replication abilities of the virus. The main signs and symptoms of CA6 strain TW-2007-00141 induced HFMD were fever and herpes appearing, but the parental virus induced HFMD caused serious symptoms, such as nail shedding, or nail loss. It remained to be further investigated whether the difference of amino acid in the 5’ UTR and 3D protein region of CA6 strain TW-2007-00141 and parental virus were related to the different clinical manifestations of HFMD caused by the two viruses. In this study, we constructed the infectious cDNA clone of CVA6 virus isolated from a patient with nail shedding, which laid the foundation for further investigation of pathogenesis of CVA6 virus induced nail shedding.
Conclusions
In this study, we successfully constructed a reverse genetic system for CVA6. After restriction enzyme digestion and immunological identification, CVA6 was successfully rescued and designated CVA6-89R. The phenotypic changes in the rescued virus were the effects of gene manipulation. The establishment of a reverse genetic system for CVA6 laid the foundation for further investigation of its pathogenic mechanism, and the dynamics of its spread, and the construction of an animal model of CVA6-induced HFMD.
Acknowledgements
This work was supported by Public Interest Technology Application Research Project, Research Plan No 2015C37124; Science and Technology Department of Zhejiang Province, China.
Conflict of Interest
The authors declare no conflict of interest.
References
- Aswathyraj S, Arunkumar G, Alidjinou EK, Hober D. Hand, foot and mouth disease (HFMD): emerging epidemiology and the need for a vaccine strategy. Med Microbiol Immunol 2016; 205: 397-407.
- Repass GL, Palmer WC, Stancampiano FF. Hand, foot, and mouth disease: identifying and managing an acute viral syndrome. Cleveland Clin J Med 2014; 81: 537-543.
- Goksugur N, Goksugur S. Images in clinical medicine. Hand, foot, and mouth disease. N Engl J Med 2010; 362: 49.
- Koh WM, Bogich T, Siegel K, Jin J, Chong EY, Tan CY, Chen MI, Horby P, Cook AR. The epidemiology of hand, foot and mouth disease in Asia: a systematic review and analysis. Pediatr Infect Dis J 2016; 35: 285.
- Wang P, Goggins WB, Chan EY. Hand, foot and mouth disease in Hong Kong: a time-series analysis on its relationship with weather. Plos One 2016; 11: 0161006.
- Yan W, Yeo A, Phoon MC, Tan EL, Poh CL, Quak SH, Chow VT. The largest outbreak of hand; foot and mouth disease in Singapore in 2008: the role of enterovirus 71 and coxsackievirus A strains. Int J Infect Dis 2010; 14; 1076.
- Yang B, Lau EHY, Peng W, Cowling BJ. Transmission of hand, foot and mouth disease and its potential driving factors in Hong Kong. Sci Rep 2016; 6: 27500.
- Liu W, Wu S, Xiong Y, Li T, Wen Z, Yan M, Qin K, Liu Y, Wu J. Co-circulation and genomic recombination of coxsackievirus A16 and enterovirus 71 during a large outbreak of hand, foot, and mouth disease in Central China. Plos One 2014; 9: 96051.
- Teng S, Zhao SY, Wei Y, Shao QM, Jiang MY, Cui DW, Xie GL. Observation on virus shedding periods of enterovirus-71 and coxsackievirus A 16 monitored by nucleic acids determination in stool samples of children with hand, foot and mouth disease. Zhonghua Er Ke Za Zhi Chinese J Pediatr 2013; 51: 787.
- Zhou HT, Guo YH, Chen MJ, Pan YX, Xue L, Wang B, Tao SH, Yu N. Changes in enterovirus serotype constituent ratios altered the clinical features of infected children in Guangdong Province, China, from 2010 to 2013. BMC Infect Dis 2016; 16: 399.
- Bracho MA, Fernando GC, Ana V, Juan C, Antonio S. Enterovirus co-infections and onychomadesis after hand, foot, and mouth disease, Spain, 2008. Emerg Infect Dis 2011; 17: 2223-2231.
- Fujimoto T, Iizuka S, Enomoto M, Abe K, Yamashita K, Hanaoka N, Okabe N, Yoshida H, Yasui Y, Kobayashi M, Fujii Y, Tanaka H, Yamamoto M, Shimizu H. Hand, foot, and mouth disease caused by coxsackievirus A6, Japan, 2011. Emerg Infect Dis 2012; 18: 337-339.
- Puenpa J, Chieochansin T, Linsuwanon P, Korkong S, Thongkomplew S, Vichaiwattana P, Theamboonlers A, Poovorawan Y. Hand, foot, and mouth disease caused by coxsackievirus A6, Thailand, 2012. Emerg Infect Dis 2013; 19: 641-643.
- Sun B, Xu S, Cui W, Liu H, Sun Y, Wu J. The analysis of the etiological surveillance results of Hand-foo-mouth disease in Changchun in 2013. Zhongguo Shiyan Zhenduanxue 2015; 5: 762-764.
- Li JL, Yuan J, Yang F, Wu ZQ, Hu YF, Xue Y, Zhou BP, Jin Q. Epidemic characteristics of hand, foot, and mouth disease in southern China, 2013: coxsackievirus A6 has emerged as the predominant causative agent. J Infect 2014; 69: 299.
- Guan H, Wang J, Wang C, Yang M, Liu L, Yang G, Ma X. Etiology of multiple non-EV71 and non-CVA16 enteroviruses associated with hand, foot and mouth disease in Jinan, China, 2009-June 2013. Plos One 2015; 10: 0142733.
- Hongyan G, Chengjie M, Qiaozhi Y, Wenhao H, Juan L, Lin P, Yanli X, Hongshan W, Xingwang L. Hand, foot and mouth disease caused by coxsackievirus A6, Beijing, 2013. Pediatr Infect Dis J 2014; 33: 1302-1303.
- Hu YQ, Xie GC, Li DD, Pang LL, Xie J, Wang P, Chen Y, Yang J, Cheng WX, Zhang Q, Jin Y, Duan ZJ. Prevalence of coxsackievirus A6 and enterovirus 71 in hand, foot, and mouth disease in Nanjing, China in 2013. Pediatr Infect Dis J 2015; 34: 951-957.
- Lu J, Zeng H, Zheng H, Yi L, Guo X, Liu L, Sun L, Tan X, Li H, Ke C, Lin J. Hand, foot and mouth disease in Guangdong, China, in 2013: new trends in the continuing epidemic. Clin Microbiol Infect 2014; 20: 0442.
- Ramirezfort MK, Downing C, Doan HQ, Benoist F, Oberste MS, Khan F, Tyring SK. Coxsackievirus A6 associated hand, foot and mouth disease in adults: clinical presentation and review of the literature. J Clin Virol Off Publ Pan Am Soc Clin Virol 2014; 60: 381-386.
- Zhang L, Wang X, Zhang Y, Gong L, Mao H, Feng C, Ojcius DM, Yan J. Rapid and sensitive identification of RNA from the emerging pathogen, coxsackievirus A6. Virol J 2012; 9: 298.
- Zeng H, Lu J, Zheng H, Yi L, Guo X, Liu L, Rutherford S, Sun L, Tan X, Li H, Ke C, Lin J. The epidemiological study of coxsackievirus A6 revealing hand, foot and mouth disease epidemic patterns in Guangdong, China. Sci Rep 2015; 5: 10550.
- Yamayoshi S, Iizuka S, Yamashita T, Minagawa H, Mizuta K, Okamoto M, Nishimura H, Sanjoh K, Katsushima N, Itagaki T, Nagai Y, Fujii K, Koike S. Human SCARB2-dependent infection by coxsackievirus A7, A14, and A16 and enterovirus 71. J Virol 2012; 86: 5686.
- Buttery VW, Kenyon C, Grunewald S, Oberste MS, Nix WA. Atypical presentations of hand, foot, and mouth disease caused by coxsackievirus A6-Minnesota, 2014. MMWR Morbid Mortal Weekly Rep 2015; 64: 805.
- Chang LY, Lee CY, Kao CL, Fang TY, Lu CY, Lee PI, Huang LM. Hand, foot and mouth disease complicated with central nervous system involvement in Taiwan in 1980-1981. J Formosan Med Assoc 2007; 106: 173-176.
- Disease H. Notes from the field: severe hand, foot, and mouth disease associated with coxsackievirus A6-Alabama, Connecticut, California, and Nevada, November 2011-February 2012. Morbid Mortal Weekly Rep 2012; 61: 213.
- Racaniello VR, Baltimore D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 1981; 214: 916-919.
- Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Nat Acad Sci 2000; 97: 6108.
- Lei Z, Wang X, Zhang Y, Gong L, Mao H, Feng C, Ojcius DM, Yan J. Rapid and sensitive identification of RNA from the emerging pathogen, coxsackievirus A6. Virol J 2012; 9: 1-6.