ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 14
A novel method for enhancing the classification of pulmonary data sets using generative adversarial networks
1Department of Computer, Lobachevsky University, Nizhny Novgorod, Russia
2Department of Computer Engineering, Ferdowsi University of Mashhad, Artificial Intelligence, Mashhad, Iran
- *Corresponding Author:
- Nasibeh Esmaeilishahmirzadi
Department of Computer
Lobachevsky University
Nizhny Novgorod, Russia
Accepted date: July 26, 2018
DOI: 10.4066/biomedicalresearch.29-18-798
Visit for more related articles at Biomedical ResearchCurrently, the use of deep neural networks in the field of processing medical images is increasing. Particularly, these networks are widely used in medical applications such as medical diagnosis. Recently, the use of generative adversarial networks has been increasing in various applications. In this paper, we propose a new method for improving and classifying a CT scan dataset of a lung image based on generative adversarial networks. Images are selected from LUNA16 dataset. After pre-processing the dataset of images and selecting the candidate areas, we divided nodule images into 3 groups of small, medium and large, and configured a generative adversarial network. Then we applied a dataset of nodule image in 3 categories, along with a random normal vector as inputs of the network. By using this method, equally we increased our dataset of images in both class of nodule and non-nodule images. Finally, we use 6 types of the CNN neural network as feature extraction and classifier on a dataset of new generated images. Compared to the use of data augmentation and use of pre-trained networks and fine-tuning, the high accuracy of the proposed method demonstrates its optimal efficiency.
Keywords
Lung images, Pulmonary nodules, Generative adversarial networks, Convolutional neural networks.
Introduction
Medical images play an important role in medicine and can contain useful information for diagnosis of diseases, monitoring treatment responses and disease management of patients with faster speed. All governments have come to the conclusion that the data produced in hospitals and insurance companies should be processed in order to help the human health.
Lung cancer is leading cause of cancer death in both men and women in the United States, accounting for 27% of cancer deaths in 2014 [1]. The challenge in the lung images is to determine and separate tumor from other part of the lung. Also in medical images de-noising and enhancement of a picture are main factors required [2]. Detection of lung nodule is essential before starting its treatment.
In the last several years computer vision also has made huge strides, mostly due to deep learning. The comparison between deep learning algorithms and shallow learning architecture shows better performance of deep learning in pattern recognition and feature extraction.
In this paper we present a technique to classification of pulmonary nodules by using convolutional neural network. Today, having an intelligent diagnostic system in order to help expert physician should be considered as a major contribution to entrepreneurship in the medical fields. Using this project in the industry, can help doctors to diagnose nodules. Image classification is the process of dividing an image into two or more number of groups. The image classification helps us to have a more detailed analysis of the task.
We have classified images using deep learning methods. The database used in this paper is derived from the largest reference database available for pulmonary nodules, called LIDC-IDRI [3].
The outline of this paper is as follows. In section 2, provide the literature review. In section 3, provide explanations on adversarial generative networks. In section 4, is explained the data set and provide the proposed method. Finally, discussion and conclusion are given in the sections 5 and 6.
Literature Review
In 2006, Hinton et al. [4] proposed pre-training and fine-tuning strategies in order to to effectively learn deep learning and created the first deep learning systems. In the last few years, various deep learning models have been developed. The most common types of deep learning models are stacked auto encoder [5-7], deep belief networks [8], convolutional networks [9] and recursive neural networks [10].
Recently, the use of deep neural networks, in particular convolutional neural networks, has grown further in medical diagnostics [11]. In 2015, Roth et al. [12] have been using convolutional neural networks to detect spleen sclerotic metastasis, lymph nodes, and colon polyps. Their research has shown that neural networks have good ability for using in CADe systems. They also increased the sensitivity and recall rates between 15% and 30% on each issue. In 2016, Setio et al. [13] proposed a new computer diagnostic (CAD) system for pulmonary nodules using multi-dimensional convolutional networks that learn the discriminative features from training data automatically. The sensitivity of this system is 85.4% and 90.1%, respectively, in 1 and 4 positive errors per scan. In the same year, Sun et al. [14] were designed deep learning algorithms and implemented, including Convolutional Neural Network (CNN), Deep Belief Networks (DBNs), Stacked Denoising Autoencoder (SDAE) and compared traditional methods of computer diagnosis and feature learning on the LIDC-IDRI data set for the purpose of classifying pulmonary nodules. The result of their research was accuracy of 0.797 in convolutional neural networks, which increased slightly compared with the traditional method with a accuracy of 0.7940. Anthimopoulos et al. [15] used convolution neural networks to evaluate and classify lung disease patterns. Their proposed network consists of 5 convolutional layers with a 2 × 2 kernel and the LeakyReLU activity function. The accuracy of 0.8561 shows the success of convolutional neural networks in classifying pulmonary patterns.
Methodology
Generative adversarial network
So far, the most striking successes in deep learning have involved discriminative models, usually those that map a highdimensional, rich sensory input to a class label [16,17]. These striking successes have primarily been based on the backpropagation and dropout algorithms, using piecewise linear units [18-20] which have a particularly well-behaved gradient. Deep generative models have had less of an impact, due to the difficulty of approximating many intractable probabilistic computations that arise in maximum likelihood estimation and related strategies, and due to difficulty of leveraging the benefits of piecewise linear units in the generative context. The generative model estimation procedure that sidesteps these difficulties. In the adversarial nets framework, the generative model is pitted against an adversary.
A discriminative model that learns to determine whether a sample is from the model distribution or the data distribution. The generative model can be thought of as analogous to a team of counterfeiters, trying to produce fake currency and use it without detection, while the discriminative model is analogous to the police, trying to detect the counterfeit currency. Competition in this game drives both teams to improve their methods until the counterfeits are in distiguishable from the genuine articles.
The adversarial modeling framework is most straightforward to apply when the models are both multilayer perceptions. To learn the generator’s distribution p g over data x, we define a prior on input noise variables pz (z) then represent a mapping to data space as G (z; θ g), where G is a differentiable function represented by a multilayer perceptron with parameters θ g. We also define a second multilayer perceptron D (x; θ d) that outputs a single scalar. D (x) represents the probability that x came from the data rather than p g. We train D to maximize the probability of assigning the correct label to both training examples and samples from G. We simultaneously train G to minimize log (1-D (G(z))). In other words, D and G play the following two-player minimax game with value function V (G, D).

The generator G implicitly defines the distribution of the probability p g as the sample distribution, which G (z) is obtained when z ~ pz [21].
Experimental Result
Dataset
The data set used in this work was collected from the largest publicly available reference database for lung nodules: the LIDC-IDRI [3].
We also used the patient lung CT scan dataset with labeled nodules from the Lung Nodule Analysis 2016 (LUNA16) Challenge. The LUNA16 [22] database are selected from the publicly available LIDC/IDRI database, and consist of 888 3D CT volumes and were provided as MetaImage (.mhd) images that can be accessed and downloaded from the LUNA16 website [23].
The CT scans from Luna 16 dataset consist of 512 × 512 slices. The annotation files in Csv format and contains candidate area of images and nodules. Also these annotation files are provided the diameter of nodules and corresponding x, y, and z locations of nodules and non-nodules in the scans. The nodule<3 are omitted and only the nodules>3 are considered as meaningful nodules.
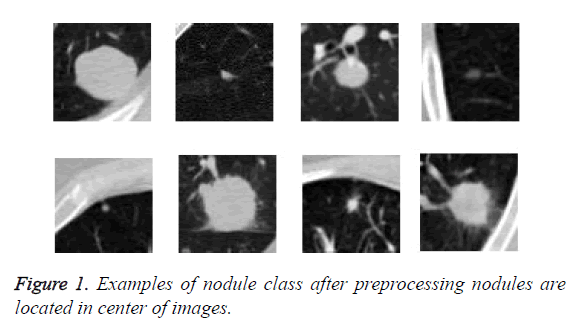
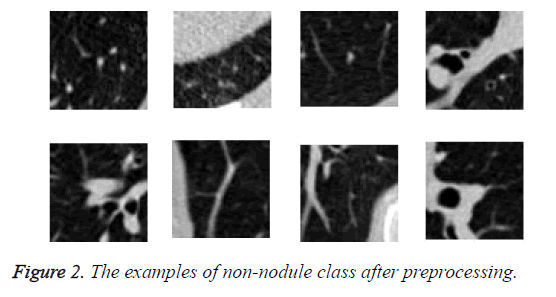
We read the raw scan by using the Simple Insight Segmentation and Registration Toolkit (SimpleITK) [24], and converted it into an array. We also read the annotations files in Csv format. The candidate locations in annotation files in world coordinates so are converted to non-integer voxel coordinates. Then the images were normalized and cropped around the candidate nodule to generate a 64 × 64 Jpg files and save them in nodule and non-nodule folders. As we can see in Figure 1 nodules are located in center of images. The diameter of nodules in range of 3.253443196 mm to 32.27003025 mm. The examples of class of non-nodule after preprocessing are depicted in Figure 2.
Totally, obtained 1005 nodules and 547680 non-nodules. Therefore, we do not need to work with the whole image, but with pieces of the image in which have the necessary information.
Proposed method
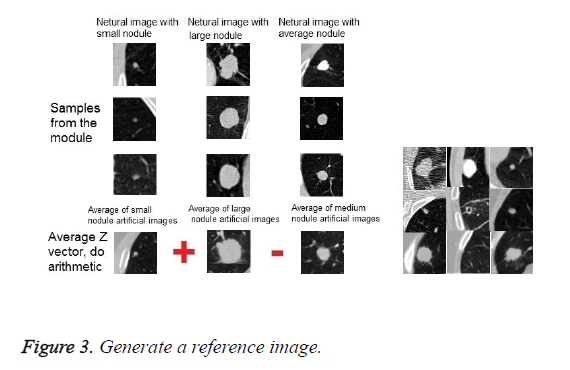
The proposed method is performed on the obtained data after the preprocessing. First, we divide the data sets of nodules into three groups of small nodules, medium nodules and large nodules. This classification is based on the diameter of the nodules. Then we configured a generative adversarial network. Subsequently, we randomly performed the complete set of nodules, including small, medium and large, with a random normal distribution vector as an input, in order to learn network to generate fake images similar to nodules and generator win in the competition between generator and discriminator. After the training of the network were completed, the network is ready to receive an image with a random normal distribution vector and generate fake images similar based on nodule or non-nodule class of images. We applied a set of nodule images in 3 categories, along with a random normal vector to the network as input. Obviously, the results of the set of fake nodule images are in 3 groups of small, medium and large. Then, we were found the mean among all fake small nodule images, and we called them as mean fake small nodules.
Similarly, we were found the mean for fake medium nodule images, and we called it as a mean of fake medium nodule images. The same procedure was performed for large nodules, which we called the mean of fake large nodule images.
Then, according to a specific relationship between these three mean images, a reference image was obtained. The exact relationship is described below. The mean of fake large nodule images plus the mean of fake small nodule images, and the obtained result is deducted from the mean of fake middle images.
We applied as input to the system, the obtained reference image, at each time which generates fake images using the generative, with the normal random vector and the actual image. In accordance with the result or output of the generative network, were obtained a fake nodule images or nodule-like images (fake images obtained from real non-nodule images).
Now, can use this generative network with the reference image, and were obtained fake nodule-like images by passing through the entire set of non-nodule class of images. This process was performed for nodule images rotationally. Finally, were generated 547680 fake nodule image and 547680 non-nodule images (nodule-like) in two classes.
Then, were used a variety of convolutional neural networks in order to classify the fake image dataset and train on it. In a test step, also did the same process and used the trained generative network in order to generate fake images by using the reference image, real images and the random normal vector.
If the input images come from class of nodule image, so the generated image will be like the actual image which inherited features of three groups of small, medium and large nodules. If the input image is class of non-nodule, so generated image will be nodule-like. Finally, by using the convolutional neural networks in order to feature extraction and classifier were obtained the corresponded class of actual images as a response.
70% of the images were used as a training image and 30% of the images were used as test image.
In this paper, we used a variety of convolutional neural networks as a classifier and feature extractor on three sets of different images in two classes. Also, we have compared the accuracy of these convolutional neural networks. We used 6 different convolutional neural networks, such as AlexNet, GoogleNet, ENet, ResNet-18, and more.
The first set of collections consists of actual images in two classes, nodule and non-nodule. The size of nodule images is 1005 and non-nodule is also 1005. The different types of network were trained weighted on this dataset with pre-training method. The second dataset consists of actual dataset of both classes with reinforcement of data in class of nodules. The size of nodule and non-nodule class is the same and equal to 547680 images, which as mentioned the size of images in nodule class was increased by using data augmentation method. In this dataset, the convolutional neural networks are fully trained.
The third dataset consists of fake images obtained from the proposed method in two classes. The size of nodule class is 547680 images and non-nodule is 547680 images. In this dataset also, the convolutional neural networks are fully trained.
The results of the classification of these three datasets of different images using different types of convolutional neural networks are shown in Tables 1-3.
| Model | Accuracy | Model | Accuracy |
|---|---|---|---|
| AlexNet | 65.23% | ResNet-34 | 75.14% |
| BN-AlexNet | 68.25% | ResNet-50 | 78.33% |
| BN-NIN | 70.21% | ResNet-101 | 80.27% |
| ENet | 71.29% | ResNet-152 | 79.59% |
| GoogleNet | 73.23% | Inception-v3 | 82.38% |
| ResNet-18 | 73.55% | Inception-v4 | 84.19% |
| VGG-16 | 74.31% | DenseNet | 85.15% |
| VGG-19 | 73.72% |
Table 1: Accuracy calculated using pre-trained networks and finetuning.
| Model | Accuracy | Model | Accuracy |
|---|---|---|---|
| AlexNet | 67.12% | ResNet-34 | 77.02% |
| BN-AlexNet | 68.59% | ResNet-50 | 80.12% |
| BN-NIN | 70.21% | ResNet-101 | 79.11% |
| ENet | 72.54% | ResNet-152 | 83.54% |
| GoogleNet | 69.12% | Inception-v3 | 84.02% |
| ResNet-18 | 70.81% | Inception-v4 | 86.11% |
| VGG-16 | 73.41% | DenesNet | 87.31% |
| VGG-19 | 75.61% |
Table 2: Accuracy calculated using data augmentation.
| Model | Accuracy | Model | Accuracy |
|---|---|---|---|
| AlexNet | 78.21% | ResNet-34 | 90% |
| BN-AlexNet | 80.32% | ResNet-50 | 92.30% |
| BN-NIN | 83.23% | ResNet-101 | 93% |
| ENet | 85.67% | ResNet-152 | 93.50% |
| GoogleNet | 84.31% | Inception-v3 | 94.51% |
| ResNet-18 | 84.59% | Inception-v4 | 94.32% |
| VGG-16 | 87.11% | DenseNet | 95.13% |
| VGG-19 | 86.23% |
Table 3: Accuracy calculated using the proposed method.
As shown in Table 3, the result of the proposed method, the accuracy of classification for all types of convolutional neural networks has increased. Further details and discussion of the accuracy of the classification of different networks on these three datasets will be presented in the next section.
Discussion
In this section, we discuss the advantages of our approach. Data augmentation is a way to reduce over fitting on models, where increase the amount of training data using information only in our training data [25]. One of the ways in data augmentation is GAN. In a typical GAN system, a random normal vector is used along with input data. The principle of the innovation of our proposed system is that we did not use simple GAN with a random vector, but we use the normal distribution vector, which we found as part of the research. This vector is the normal distribution of the weighted average of the data, and we apply algebraic operations between them, as shown in Figure 3. Thus, we have been developing new images that have the feature of all three small, large and medium nodules. To find this vector of normal distribution, we inherited the characteristics of small, medium, and large nodules, and we apply it as a vector of normal distribution to the GAN system.
Therefore, instead of producing fake images which are less similar to the original data, the GAN system using the proposed method, can produce images, which are more similar to the original images since data processing has been applied to the original data characteristics. In other words, during the production of the data we limited the GAN in order to inheritance from the main data feature.
The advantage of the proposed model is that it affects the accuracy of the classification and improves it.
As shown in the Table 1, the accuracy of the classification of all kinds of convolutional neural network in the proposed method, according to the training on the dataset of generated images, are better than the other datasets in Tables 1 and 2. Table 1 relates to the training of all types of convolutional neural networks on an actual dataset of pre-trained, and Table 2 relates to the training of all types of convolutional neural networks on a dataset of images, with the reinforcement of a nodule class by data augmentation method, and Table 3 relates to training of convolutional neural networks on the collection of fake images produced by the proposed method.
The disadvantages of the proposed system is to impose an additional computational load and time-consuming on the system. In general, it can be concluded that the data augmentation adds additional load to the convolutional networks in terms of time and computation, but does not have much effect on the testing stage.
Conclusion
Lung cancer is one of the causes of cancer death around the world. Early diagnosis of pulmonary nodules helps to diagnose the disease and prevent death from the disease. The diagnosis of pulmonary diseases by the physician requires a lot of time and attention. To reduce the time and increase the accuracy and efficiency, as well as accelerate the diagnosis, computer diagnostic systems have been developed with the help of physicians to reduce the death rate of the diseases.
Our challenge in classification of lung nodules is very sensitive because in medical images, the number of nodules is fewer than the non-nodules. So the network tends to detect of non-nodule images, and it is less likely to detect nodule images. This challenge leads us to weight training and testing to make the network sensible to the diagnosis of nodule images. Images are taken from LUNA 16 Dataset, which is derived of the LIDC-IDRI database.
Due to the fact that the ratio of nodule images to non-nodules is unbalanced, either needs the data augmentation to increase the nodule images or use weighted training. Otherwise, the accuracy of detection is not desirable and consequently, it does not acceptable results in the real world.
If we suppose that the network detects all the nodule images in the dataset of images as non-nodule, so, due to the imbalance between the number of images in two classes and the multiplicity of the number of non-nodule class of images relative to the number of nodule class of images, the system's accuracy is higher than 90% and this result is inacceptable.
Two factors improve the accuracy of classification in the proposed method. First using the generative adversarial network in order to generate fake images based on 3 groups of nodules combinatorially and inherits feature of small, middle and large nodules, which has led to the dataset of nodule images consist of images which are similar to combination of these three group. Also the dataset of fake non-nodule images consist of images which tend to be like these images and we called them nodule-like images. The considerable number images of fake images in two classes led to the optimal training of various kinds of convolutional neural network as a feature extractor and classifier.
The second factor of advantage of the proposed method is the uniformity of the training and test data. As mentioned before, the training set of data in the proposed method is artificially generated. During the test, any image by passing the generative network, is converted to a fake image, and apply in artificial neural network classifier for final detection. This factor also improves the classification efficiency.
References
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin 2014; 64: 9-29.
- Waseem K. Image segmentation techniques: a survey. J Image Graphics 2013; 1: 4.
- Armato Samuel G. The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 2011; 38: 915-931.
- Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science 2006; 313: 504-507.
- Gehring J, Miao Y, Metze F, Waibel A. Extracting deep bottleneck features using stacked auto-encoders. Proc IEEE Int Conf Acoust Speech Sig Proc 2013; 26-31.
- Weng R, Lu J, Tan Y, Zhou J. Learning cascaded deep auto-encoder networks for face alignment. IEEE Trans Multimedia 2016; 18: 2066-2078.
- Sun M, Zhang X, Hmme HV, Zheng TF. Unseen noise estimation using separable deep auto encoder for speech enhancement. IEEE/ACM Trans Audio Speech Lang Process 2016; 24: 93-104.
- Hinton GE, Salakhutdinov RR. Reducing the dimensionality of data with neural networks. Science 2006; 313: 504-507.
- Karpathy A, Toderici G, Shetty S, Leung T, Sukthankar R, Li F. Large-scale video classification with convolutional neural networks. Proceedings of IEEE Conference on Computer Vision and Pattern Recognition 2014; 1725-1732.
- Gers FA, Schraudolph NN, Schmidhuber J. Learning precise timing with ISTM recurrent networks. J Mach Learn Res 2002; 3: 115-143.
- LeCun Y, Koray K, Clement F. Convolutional networks and applications in vision. Circuits and Systems (ISCAS), Proceedings of 2010 IEEE International Symposium 2010.
- Roth Holger R. Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imag 2016; 35: 1170-1181.
- Setio AAA. Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imag 2016; 35: 1160-1169.
- Sun W, Bin Z, Wei Q. Computer aided lung cancer diagnosis with deep learning algorithms. Med Imag Comp Aid Diag Int Soc Optic Photon 2016; 9785.
- Anthimopoulos M. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imag 2016; 35: 1207-1216.
- Krizhevsky A, Sutskever I, Hinton G. ImageNet classification with deep convolutional neural networks. NIPS 2012.
- Hinton G, Deng L, Dahl GE, Mohamed A, Jaitly N, Senior A, Vanhoucke V, Nguyen P, Sainath T, Kingsbury B. Deep neural networks for acoustic modeling in speech recognition. IEEE Sig Proc Magaz 2012; 29: 82-97.
- Goodfellow IJ, Warde-Farley D, Mirza M, Courville A, Bengio Y. Maxout networks. ICML 2013.
- Glorot X, Bordes A, Bengio Y. Deep sparse rectifier neural networks. AISTATS 2011.
- Jarrett K, Kavukcuoglu K, Ranzato M, LeCun Y. What is the best multi-stage architecture for object recognition? Proc IEEE International Conference on Computer Vision (ICCV09) 2009; 2146-2153.
- Goodfellow I. Generative adversarial nets. Adv Neur Info Proc Sys 2014.
- Setio AAA. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: the LUNA16 challenge. Med Image Anal 2017; 42: 1-13.
- https: //luna16.grand-challenge.org/download/
- Lowekamp BC, Chen DT, Ibanez L, Blezek D. The design of simple ITK. Front Neuroinform 2013; 7: 45.
- Perez L, Wang J. The effectiveness of data augmentation in image classification using deep learning. Comp Vis Patt Recogn 2017.