ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 21
Starvation contributes to elevated levels of heat shock proteins and cancer stem cell markers in an esophageal cancer cell line.
Amer Talib Tawfeeq*, Noah Abd-Alkader Mahmood, Zaynab Saad Abd-Alghni
Department of Molecular Biology, Molecular Biology Lab, Iraqi Center for Cancer and Medical Genetics Research, Iraq
- *Corresponding Author:
- Amer Talib Tawfeeq
Department of Molecular Biology
Molecular Biology Lab
Iraqi Center for Cancer and Medical Genetics Research
Iraq
Accepted date: November 26, 2018
DOI: 10.4066/biomedicalresearch.29-18-1138
Visit for more related articles at Biomedical ResearchThe presence of cancer stem cells as a subpopulation residing in the apex of solid tumor cell hierarchy has been introduced as a new hypothesis capable of describing the ability of tumors to be more aggressive, highly metastasized, and chemo-radiotherapy resistant. The origin of these cells is still controversial: one hypothesis describes the stress conditions in the tumor microenvironment as one of the driving forces behind the existence of these cells. In this study, we test if nutrition depletion conditions (deprivation of serum and glucose) would be one of such forces. Esophagus adenocarcinoma cell line SKGT-4 was exposed to starvation by depleting glucose and fetal bovine serum from growth media at different times. The viability of the cells during this condition was determined by standard MTT assay and the cells’ morphological changes were observed by crystal violet staining and trypan blue staining. The expression levels of stress-related proteins, heat shock protein 90 (HSP90) and heat shock protein 70 (HSP70), as well as some known cancer stem cell markers, CD44, ALDH1A1, and ABCG2, were determined using quantitative real-time PCR. Levels of necrosis and apoptosis were followed in cell populations under stress using a mixture of fluorescence staining and observation under a fluorescence microscope. The results indicated a loss of cell viability during the extended times of incubation in starved condition compared with the non-starved condition. Cells under starvation suffered from noticeable morphological changes combined with widespread necrosis and apoptosis. Levels of HSP90, HSP70, and cancer stem cell marker expression were significantly increased in starved condition compared with non-starved condition (p ≤ 0.01). In conclusion, although starvation as a result of serum and glucose depletion leads to induced necrosis and/or apoptosis in most of the cells, it may induce stress-resistant mechanisms in cells that remain viable (stress-resistant cells).
Keywords
Cancer stem cells, Starvation stress, Cancer microenvironment, Heat shock proteins, CD44
Introduction
Cancer stem cell hypothesis describes a subset of cells responsible for tumor initiation and maintenance [1]; their rise from normal stem cells or other cells of the tumor remains to be elucidated. These cells contribute to major hallmarks of cancer; they garner much attention since they make this disease harder to cure [2].
Based on the cancer stem cell concept, tumor cell heterogeneity is attributed not only to the differences between these cell sub-clones (as a result of chaotic mutation events) but also results from tumor-resident CSC/progenitor cell types [3-7].
The correlation between cancer stem cells and stresses they encounter in tumor microenvironments such as hypoxia, serum starvation, and hyperthermia, is unclear [8,9]. Such stresses attribute to cancer cell death, plasticity, and epithelialmesenchymal transition [10]. In recent years, researchers used hypoxic incubation conditions, serum-free media, and elevated incubation temperatures on cell line models to simulate the stress imposed by tumor microenvironment in order to understand its effects on cancer cell stress, plasticity, and epithelial-mesenchymal transition [9,11,12].
It is well-established that heat shock proteins such as HSP70 and HSP90 play a substantial role in cancer cell resistance to stress, thereby elevating tumor invasiveness, relapse, and drugresistance [13]. These molecular chaperones were found to be highly expressed in different types of cancer cells: they play a protein chaperoning role intensively during stressful conditions [14].
The World Health Organization reports that more than 76,000 deaths were registered as a result of esophageal cancer [15]. This type of cancer can be difficult to treat and becomes resistant to currently available standard therapies, which may be attributed to the prevalence of cancer stem cells in this type of tumor. Therefore, intensive research has been started to study the types of CSC markers and understand their critical role in the biology of esophageal epithelium disease [16]. In accordance with many types of cancers, most comprehensive esophagus CSC markers that were recognized included CD44, ABCG2, and ALDH, besides other minor markers [16,17].
In this research, we asked if the starvation conditions that cancer cells encounter in the tumor microenvironment could be a driving force for the enrichment of the cancer stem cell subpopulation. Furthermore, we hypothesized if there is any correlation between HSP70 and HSP90 levels and the level of some cancer stem cell markers such as CD44, ALDH, and ABCG2 during nutrition depletion represented by serum starvation and glucose deprivation throughout extended incubation periods in the esophageal cancer cell line (SKGT4).
Materials and Methods
Cell line and cultures
The esophageal adenocarcinoma cancer cell line (SKGT-4) was obtained from the cell bank unit in the Iraqi Center for Cancer and Medical Genetics Research. Cells were cultured in 25 ml tissue culture vessels (Santa Cruz Biotechnology, California, USA) grown in RPMI-1640 culture media with L-glutamine and glucose (United States Biological, Salem, MA 01970, USA) and 10% fetal bovine serum (FBS) (Capricorn Scientific GmbH, Auf der Lette 13 A, 35085 Ebsdorfergrund, Germany) until they reached a confluent monolayer. After reaching the status of the confluent monolayer, the cultures were divided into two groups according to the type of incubation conditions. In the first type of incubation condition, cultures were incubated under normal growth conditions in RPMI-1640 (United States Biological, Salem, MA 01970, USA) growth media with L-glutamine, 5% glucose, and 10% fetal Bovine serum. The timeline for this type of incubation condition was 24, 48, 72, and 96 h. During these times, the cultivation media was not changed, and the cells were left to grow in the same media for the entire incubation period. In the second type of incubation condition, cultures were grown under starvation in RPMI-1640 growth media (United States Biological, Salem, MA 01970, USA) without glutamine, without glucose, and without FBS. As in the first type of incubation condition, the cultivation media was not changed and the cells were left to grow in the same media for the entire incubation period. Both types of incubation condition used growth media containing 1% penicillin/streptomycin and were incubated at 37°C in a humidified atmosphere containing 5% CO2 for 24, 48, 72, and 96 h (Figure 1).
Figure 1: The design of the serum and glucose starvation experiments, a confluent monolayer of SGKT-4 cells grown in tissue culture vessels incubated for normal non-starvation and serumglucose starvation conditions for the indicated time at 37°C, 5% CO2, and 96% humidity. The parameters detected are as indicated.
MTT assay
A standard MTT assay using 96 well plates was carried out to a determined reduction in cell viability during both types of incubation conditions (starved and non-starved cultures) for the incubation times mentioned earlier, according to the method described by Mosmann.
Crystal violet staining
Cells (1 × 105 cell/ml) were seeded in the 96 well plates in RPMI-1640 growth media (United States Biological, Salem, MA 01970, USA) with L-glutamine, 5% glucose, 10% FBS and 1% penicillin/streptomycin and incubated at 37°C in a humidified atmosphere containing 5% CO2 until it reached confluent monolayer in the wells. At that time the media was discarded from the plates and replaced by either normal growth media (with L-glutamine, 5% glucose, 10% FBS and 1% penicillin/streptomycin) or the media that did not contain FBS and glucose (starvation condition media). Plates were incubated at 37°C in a humidified atmosphere containing 5% CO2 for 24, 48, 72, and 96 h. At the end of incubation time in both types of incubation conditions, cells were fixed using cold methanol (-20°C) for 5 min, and after that, methanol was discarded and crystal violet stain (5 mg/ml) dissolved in distilled water containing 10% buffered formalin was applied over the cells in the wells (50 μl/ well). Plates were left in the incubator for 5 min, allowing the cells to absorb the stain. The stain was discarded right after, plates were washed three times with distilled water, and then the cells were imaged using a Leica inverted microscope (DMIL LED flue, Leica Microsystems, Wetzlar, Germany) compounded with a CCD Leica DFC550 camera (Leica Microsystems, Wetzlar, Germany).
RNA extraction
Total RNA was extracted from the cells using an AccuZol™ total RNA Extraction Solution (Bioneer, DMC Hi-tech Industry Center, Sangam-dong, Mapo-Gu, Seoul, South Korea), RNA purity and concentration determined using Q5000 UV-Vis microliter spectrophotometer (Colwell, San Jose, CA 95161 USA) and its integrity was demonstrated with agarose gel electrophoresis.
Quantitative real-time PCR
For quantitative reverse transcriptase PCR (qRT-PCR), the one-step SYBR green kit was used (one-step SYBR Fast, KAPA Biosystems, USA). Primers were designed for each gene using NCBI/primer-BLAST and its specificity for each gene investigated was verified using BLAST and the UCSC genome browser using the in-silico PCR tool [18] as well as during real-time PCR by the formation of a single peak during the disassociation step (Table 1). Optimum annealing temperature was optimized using gel electrophoresis over a range of different temperatures extended from 50°C to 64°C with conventional PCR (Sure Cycler 8800 Thermal Cycler, Agilent Technologies, USA). The thermal profile consisted of 42°C for 5 min to synthesize cDNA, 95°C for 3 min to deactivate reverse transcriptase (RT), followed by 40 cycles at 94°C for 15 s, 60°C for 30 s, 59°C, and 58°C for 20 s to anneal primers (according to each gene optima), 72°C for 20 s for dsDNA extension. Disassociation curve temperatures were 95°C for 1 min, 55°C for 30 s, and 95°C for 30 s. Fold expression for each gene was determined using the 2-ΔΔCt Livak method [19] in comparison with the β-actin gene as a housekeeping gene. β-actin was chosen because of its stability and consistency in the starved and fed environments and the fact that its protein, mRNA, and gene expression are widely used to normalize biological data [20].
| Gene name | Sequence 5'-3' | Tm | Accession no. | |
|---|---|---|---|---|
| HSP70B | Forward primer | 5'-ACACGAATCCCTGCGGTAAAA-3 | 60 | NM_002154 |
| Reverse primer | 5-GCAGGCGATAAGATGGCACA-3' | 60 | ||
| HSP90B | Forward primer | 5-CGAAGACTCCACTAACCGCC-3' | 62.1 | NM_001271970 |
| Reverse primer | 5-ACTCGCTCCACAAAAGCTGA-3' | 61.1 | ||
| CD44 | Forward primer | 5-TCTGCGGGCTGCTTAGTCA-3' | 63.2 | NM_001001389 |
| Reverse primer | 5'-ACCGAACCTGGAAGAGGCT-3' | 61.2 | ||
| ALDH | Forward primer | 5-CTGCATACACTTATCACAGGT-3' | 52.8 | NM_000689 |
| Reverse primer | 5-GTAACACCTAGGGCAGGAAG-3' | 56.4 | ||
| ABCG2 | Forward primer | 5'-GGAGAGCTGTTAATTTCAG-3' | 49.9 | XM_005263355 |
| Reverse primer | 5-CACGCCTACTAAACAGAC-3' | 48.8 | ||
| β-actin | Forward primer | 5'-CCGCAAATGCTTCTAGGCG-3' | 64.1 | NM_001101 |
| Reverse primer | 5-'TGTTTTCTGCGCAAGTTAGGT-3' | 60.9 | ||
Table 1. The sequences of specific primers used for determination of HSP70, HSP90, CD44, ALDH1A1, ABCG2 and β-actin mRNA level by realtime PCR.
Trypan blue staining
The same procedure for crystal violet staining was used for trypan blue staining, but in the use of this stain the cells were not fixed and stained while they remained alive. Briefly, after the end of incubation time for the intended conditions, 100 μl of the media was aspirated from each well and 10 μl (4 mg/ml) of trypan blue stain solution was added. Stained cells were visualized and imaged using a Leica inverted microscope (DMIL LED flue, Leica Microsystems, Wetzlar, Germany) compounded with a CCD Leica DFC550 camera (Leica Microsystems, Wetzlar, Germany) [21].
Detection of apoptosis and necrosis
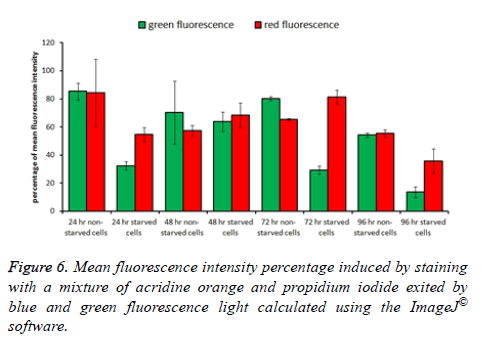
To determine the level of apoptosis and necrosis induced as a result of starvation conditions, cells were cultured in 96 flat bottom microtiter plates and exposed to starvation and nonstarvation conditions as described earlier (in crystal violet staining section) for the same incubation times. After incubation time ended in both, incubation condition media was aspirated carefully from each well and 35 μl of a mixture of acridine orange and propidium iodide fluorescence stains (0.4 mg/ml) was added to the wells. Plates were incubated for 10 min at 37°C with 96% humidity incubator, and the dye mixture was aspirated carefully from the wells at the end of incubation time. Cells were washed with 50 μl PBS one time and imaged immediately using an inverted fluorescent microscope (DMIL LED flue, Leica Microsystems, Wetzlar, Germany) compounded with a CCD Leica DFC550 camera (Leica Microsystems, Wetzlar, Germany). Using this method, both live and dead cells were characterized. Among dead cells, those which suffered apoptosis or necrosis were differentiated as well; all cell types were counted manually using 50 images for each treatment. ImageJ® software was used to calculate the total fluorescence intensity in the five images for each field of each treatment and the average was used in plotting the histogram.
Statistical analysis
The relative expression data of HSP90, HSP70 as well as cancer stem cells markers (CD44, ALDH1A1, and ABCG2) were analysed using a one-way analysis of variance (ANOVA) followed by Tukey posttest. Control cultures (non-starvation condition at 24 h) were used to compare the variation between the expression values of all parameters in the starved and nonstarved condition of the other incubation times. The probability of 95% and 99% of values were considered statistically significant and highly significant respectively. The same procedure was used in apoptosis/necrosis experiments.
Results
Cell viability
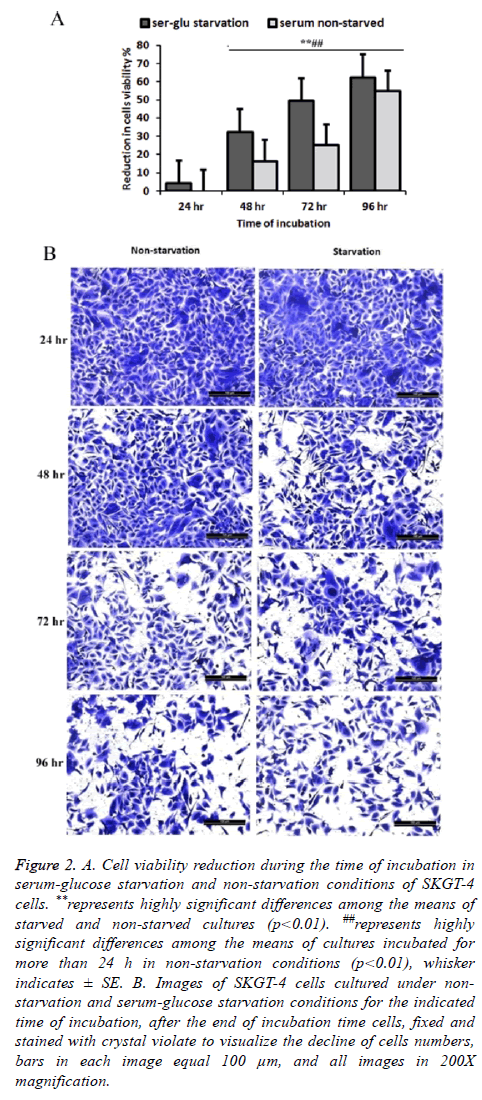
The results indicated a loss of cell viability during the extended time of incubation after cells becomes confluent monolayer on the tissue culture 96 well plates. Using the incubation time of 24 h in serum non-starvation (normal conditions) as a representative of 100% cell viability, reduction in viability of the cells for extended time incubation with and without starvation condition was calculated. There was a continuous decline of cell viability in non-starved cultures after 48, 72, and 96 h of incubation by 16.32% SE ± 1.23, 24.95% SE ± 2.04, and 54.77% SE ± 1.69 respectively. This cell viability reduction increased in starvation conditions to 4.15% SE ± 1.20, 32.16 SE ± 1.09, 49.33% SE ± 2.05, and 62.41% SE ± 2.45 at 24, 48, 72 and 96 h of the extended incubation times, respectively, in serum starvation condition (Figures 2A and 2B).
Figure 2: A. Cell viability reduction during the time of incubation in serum-glucose starvation and non-starvation conditions of SKGT-4 cells. **represents highly significant differences among the means of starved and non-starved cultures (p<0.01). ##represents highly significant differences among the means of cultures incubated for more than 24 h in non-starvation conditions (p<0.01), whisker indicates ± SE. B. Images of SKGT-4 cells cultured under nonstarvation and serum-glucose starvation conditions for the indicated time of incubation, after the end of incubation time cells, fixed and stained with crystal violate to visualize the decline of cells numbers, bars in each image equal 100 μm, and all images in 200X magnification.
Heat shock proteins and cancer stem cell markers expression in starvation and non-starvation conditions
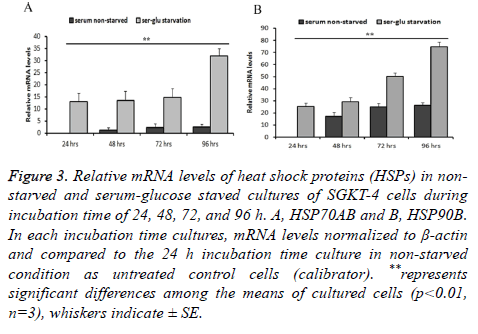
We used the 24 h culture that incubated at the non-starvation condition as a baseline expression level to compare the mRNA levels in the other culture (in both starvation and nonstarvation condition). A significant correlation between serum starvation during incubation at extended time of incubation, 72 and 96 h, and the elevated levels of mRNA for the genes of heat shock proteins (stress proteins) and cancer stem cell markers under investigation. HSP70 was found to be significantly elevated (p ≤ 0.01) at the extended time of incubation during starved cultivation conditions. Levels of HSP70 mRNA in starved cultures was significantly higher by 13.06 SE ± 3.5, 13.43 SE ± 4, 14.79 SE ± 3.6, and 32.94 SE ± 3 (n=3) times than during incubation at 24, 48, 72, and 96 h relatively to 24 h incubation time of non-starved culture.
In non-starved cultures, the gene mRNA relative level did not elevate significantly (P ≤ 0.01, n=3) (1.16 SE ± 1.2, 2.37 SE ± 1.5, and 2.56 SE ± 1) during 48, 72, and 96 h of incubation compared to the non-starved 24 h culture. The differences between starved and non-starved cultures during 48, 72, and 96 h of incubation were significant (P ≤ 0.01, n=3). It was higher in corresponding starvation conditions than in non-starvation conditions relatively by 13.06, 12.26, 12.42, and 29.38 times than in starved cultures (Figure 3A). The same was true of HSP90: this gene RNA witnessed significant elevation (P ≤ 0.01, n=3) in the starved cultures by 25.15 SE ± 2.99, 29.29 SE ± 3.56, 50.30 SE ± 2.7, and 74.67 SE ± 3.9 times than during 24, 48, 72, and 96 h of incubation. In contrary to HSP70, this gene has significantly (P ≤ 0.01, n=3) higher mRNA levels in non-starved culture during 48, 72, and 96 h of incubation (17.17 SE ± 3.55, 24.97 SE ± 3, and 26.21 SE ± 2.45) compared to 24 h nono-starved culture. The increase of mRNA in this gene was 25.15, 12.11, 25.32 and 48.45 times higher between corresponding starvation and non-starvation incubation conditions at 24, 84, 72, and 96 h respectively (Figure 3B).
Figure 3: Relative mRNA levels of heat shock proteins (HSPs) in nonstarved and serum-glucose staved cultures of SGKT-4 cells during incubation time of 24, 48, 72, and 96 h. A, HSP70AB and B, HSP90B. In each incubation time cultures, mRNA levels normalized to β-actin and compared to the 24 h incubation time culture in non-starved condition as untreated control cells (calibrator). **represents significant differences among the means of cultured cells (p<0.01, n=3), whiskers indicate ± SE.
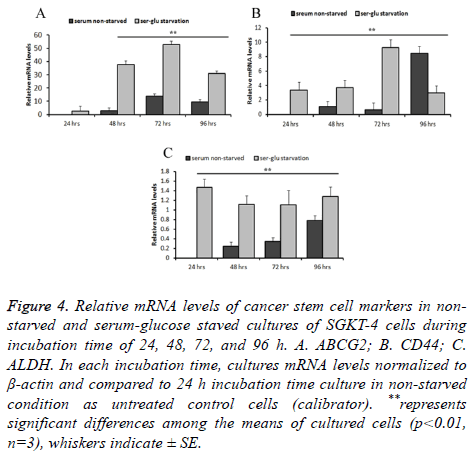
The cancer stem cell markers mRNA levels varied in a way that is more noticeable. For ABCG2, relative mRNA levels in starved cultures were not significantly (P ≤ 0.01, n=3) increased at 24 h compared to the 24 h non-starved culture (2.49 times SE ± 0.201). However, after 48, 72, and 96 h of incubation in the starvation condition the mRNA levels of this gene elevated significantly (P ≤ 0.01, n=3) by 37.46 SE ± 3.0, 52.98 SE ± 2.5, 31.07 SE ± 2 times respectively, compared to the control 24 h non-starved culture. In the non-starved cultures, incubation for extended time induced an elevation in the relative mRNA levels. It was not significant at 48 h (2.902 times SE ± 0.3) and significant at 72 and 96 h (13.71 SE ± 1.02 and 9.563 SE ± 0.93 times respectively). The effect of starvation clearly appeared in comparing levels of mRNA between starved and non-starved cultures during corresponding incubation time. It was elevated significantly (P ≤ 0.01, n=3) by 34.56, 39.27, and 21.507 times after 48, 72, and 96 h respectively (Figure 4A). Expression of CD44 was almost similar to ABCG2, its mRNA level increased significantly (P ≤ 0.01, n=3) by 3.34 SE ± 0.55, 3.71 ES ± 0.67, 9.26 SE ± 0.526 times after 24, 48, and 72 h and was decreased by 2.97 SE ± 0.49 times after 96 h in comparison with the 24 h non-starved culture. In the non-starved culture, there was no significance (P ≤ 0.01, n=3) in levels of this gene mRNA between 48 and 72 h (1.08 SE ± 0.080 and 0.654 SE ± 0.0808). Nevertheless, when incubation under non-starvation condition extended to 96 h, mRNA level increased significantly to 8.47 ES ± 0.45 times. Differences in relative mRNA level were more obvious in starved cultures in a corresponding time of incubation to the non-stared cultures at 24, 48, and 72 h. It was significantly (P ≤ 0.01, n=3) higher by 3.34, 2.62, and 8.61 times respectively. Whereas after 96 h this gene expression decreased in starved cultures by 5.49 times than in the non-starved culture (Figure 4B). Another important marker of cancer stem cells that followed was ALDH. This gene mRNA was considerably lower in both starved and non-starved cultures. However, when using the non-starved culture at 24 h as a control baseline to compare with, the mRNA relative level was much higher and significant (P ≤ 0.01, n=3) at 24, 48, 72, and 96 h. Overall, the relative level of this gene mRNA was increased in the starved cultures by 0.117, 0.085, 0.214, and 0.097 compared to nonstarved cultures at each corresponding time of incubation: 24, 48, 72, and 96 h respectively (Figure 4C).
Figure 4: Relative mRNA levels of cancer stem cell markers in nonstarved and serum-glucose staved cultures of SGKT-4 cells during incubation time of 24, 48, 72, and 96 h. A. ABCG2; B. CD44; C. ALDH. In each incubation time, cultures mRNA levels normalized to β-actin and compared to 24 h incubation time culture in non-starved condition as untreated control cells (calibrator). **represents significant differences among the means of cultured cells (p<0.01, n=3), whiskers indicate ± SE.
Cellular morphological changes
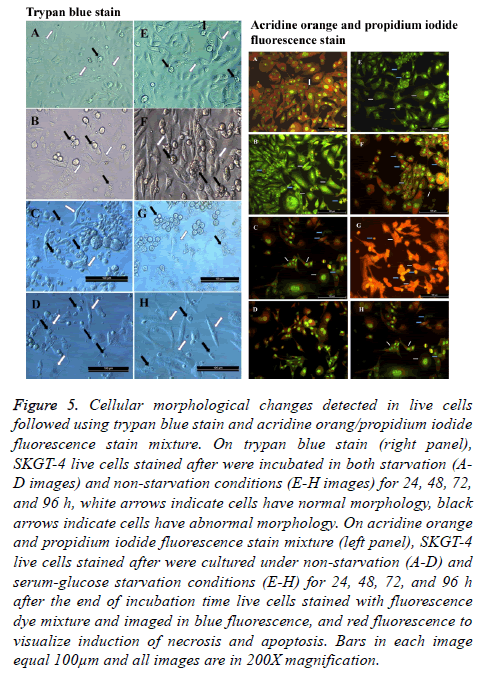
Trypan blue stain was used considerably to distinguish between live and dead cells, getting the advantage of exclusion capability of live cells toward this stain in suspension and/or adherent cultures. Whenever we have employed this stain, it was in a dual-use, first morphological changes imposed on cells were visualized under both starvation and non-starvation conditions, taking advantage of the capacity of this stain to reduce the transparency of the inspected microscopic fields as counter color. Second, these changes were observed while the cells remained live or unchanged by the effect of the fixation process. Characterizing the morphological changes, we have observed that cells under starvation suffered from massive morphological changes as seen under a light microscope. As in all experiments conducted, we tainted to concede the 24 h nonstarved cultures as the control untreated cells to compare it with the effect of starvation and extended time of incubation. During non-starvation conditions, cells continued growth and increased in number. The most noticeable morphological change was that cells started to float and aggregate. Cells took rounded shape with singes of death. Cells also suffered from plasma membrane blebbing and some of them start to shrink (Figure 5).
Figure 5: Cellular morphological changes detected in live cells followed using trypan blue stain and acridine orang/propidium iodide fluorescence stain mixture. On trypan blue stain (right panel), SKGT-4 live cells stained after were incubated in both starvation (AD images) and non-starvation conditions (E-H images) for 24, 48, 72, and 96 h, white arrows indicate cells have normal morphology, black arrows indicate cells have abnormal morphology. On acridine orange and propidium iodide fluorescence stain mixture (left panel), SKGT-4 live cells stained after were cultured under non-starvation (A-D) and serum-glucose starvation conditions (E-H) for 24, 48, 72, and 96 h after the end of incubation time live cells stained with fluorescence dye mixture and imaged in blue fluorescence, and red fluorescence to visualize induction of necrosis and apoptosis. Bars in each image equal 100μm and all images are in 200X magnification.
Detection of apoptosis and necrosis
As expected, cells under starvation and extended time of incubation suffered from both types of deaths, apoptosis, and necrosis. As seen under a fluorescent microscope with green and blue fluorescent light, the cells undergoing starvation were more apoptosis and necrosis susceptible as a result of a shortage in nutrition (Figure 5). There were cells that did not show any morphological changes and remained attached as a sign of normal living cells, that makes them exceptionally noticed in the course of events in such conditions. These cells are resistant to the conditions imposed by the surrounding conditions [22,23].
Measuring total fluorescence intensity in both types of cultures after the defending time of incubation revealed a consistency with the observed images. A cell stained with green indicates a sound live cell, while cells stained with red indicates a dead cell suffering either apoptosis or necrosis. The highest green fluorescence intensity percentage was observed in non-starved cultures incubated for 24 h and the lowest green fluorescence intensity percentage was observed in starved cultures incubated for 96 h. The remaining cultures ranged in both their green and red fluorescence intensity percentages between those two mentioned cultures (Figures 5 and 6).
Discussion
It is more than a decade now since cancer stem cell markers were first recognized in solid tumors (breast cancer). Since then huge efforts from hundreds of teams working around the world started to investigate different types of solid tumors to determine the types of markers that may be related to normal stem cell characteristics and still be found in cancer cells [24]. After establishing the kinds of cancer stem cell markers in many types of tumors, their contribution to the aggressiveness of the tumors they found was recognized. It is known that cancer stem cells markers vary among different types of cancer, but they all contribute to the major cancer hallmarks such as metastasis chemo-radiotherapy resistant and they represent a new therapeutic target opportunity [25,26]. Then the question about the origin of these cells emerged, whether they originated from mutated normal tissue-specific stem cells or maybe as a result of some kind of transition of differentiated cancer cell backward to cancer stem cell in tumor population hierarchy, non-CSC to CSC, where the term of cancer cell plasticity was introduced. This cancer cell plasticity suggested being increased when cancer cells counter stressful circumstances such as chemo-radiotherapy. Therefore the relationship between cancer stem cells that lay on the apex of tumor cell hierarchy and their progeny cancer cells may embrace bidirectional inter-conversions [27-29].
Tumor niche suffers from harsh micro-environmental stresses such as low pH, hypoxia, and inflammation because of poor vesicles. These conditions impose a kind of necessity for the cancer cell to induce certain mechanisms in order to overcome such conditions [30]. Besides these kinds of abnormal conditions, we have asked about the extent that nutrition depletion role in such conditions. Therefore we designed the experiment as if the increment of cell number associated with the lack of serum and glucose in the growing media mimic the environment of the poorly vasculature tumors. Results of this research indicated that nutrition depletion may play a critical role in changing some of the important characters in cancer cells.
We have followed what should be the most noticeable changes that could be followed with the simplest methodology available. We set out to detect the reduction in cellular viability, deformations in cellular morphology, and link those with the levels of heat shock proteins and cancer stem cell markers of the esophagus cell line. The final goal was to test the capacity of this type of cancer to induce changes in its most important characters in order to survive and as a result become more aggressive if some changes may take place in the nutrients of the surrounding environment. The results we find in this study indicate that serum and glucose starvation contribute substantially to the reduction of cells viability with massive cellular morphological changes and relate directly to the increment of cancer stem cell markers in esophageal cancer cell line SKGT-4. As primer need, to assess the differences in the levels of heat shock proteins and cancer stem cell markers expression during serum and glucose starvation through the extended time of incubation, we need a starting baseline to compare with. Therefore, we need a fully growing culture that does not suffer any kind of stress during its lifetime. To fulfil this condition, we used the confluent monolayer culture of the 50 ml standard cultivation vessel, which was kept for only 24 h with 10% FBS and 5% glucose after reaching monolayer status. The cellular viability, cellular morphological shape, and mRNA extracted from this culture were used as a reference starting baseline to compare it with the effect of starvation during extended incubation time, which in this study was for 48, 72, and 96 h. Besides that, we used this culture to compare the effect of extending incubation time of monolayer cultures grown in 96 well standard tissue culture microtiter plates in the presence of 10% FBS and 5% glucose (non-starvation conditions) on the type of cellular death, whether it is necrosis or apoptosis. It is well known that when cancer cells reach monolayer their growth will not stop and they will compete to survive since there will be a shortage in nutrients and accumulation of acidity and other byproducts [31]. These unfavorable growth conditions will induce apoptosis in the population while some other cells will struggle to keep a life. In that environment cells will induce different types of survival mechanisms; in accordance, we tested if the genes under investigation are among that survival machinery. From the results presented here, heat shock proteins 70 and 90 with other cancer stem cell markers elevated considerably during stress. This elevation was combined by a massive reduction of cells viability as a result of the induction of both mechanisms of cellular deaths apoptosis and necrosis.
What may be seen as a conflict in this result is that the decrease in cells number in cultures under investigation was a pronoun, yet there was a noticeable elevating in the level of the mRNA for the genes under study. In the RT-qPCR technique, it's mandatory to unify the quantities of the RNA purified from different samples that would be used in expression comparisons (relative expression). This will lead by definition to dilute the RNA extracted from samples with a larger cell number to the levels of that extracted from samples with a smaller cell number. Whoever, the key point here, is the amount of the RNA of the specific genes in these samples that would be determined in RT-qPCR. In our results, this situation was evidently noticed. In the cultures that suffered from starvation, the cell count was decreased as a result of apoptosis and necrosis, therefore, the total amount of RNA purified from such cultures was less than that purified from non-starved cultures. What our results indicate here is that the RNA of HSP90 and 70, as well as the RNA of cancer stem cell markers, was elevated in the total amount of the RNA investigated in starved cultures compared to that of non-starved cultures. These results indicate a higher level of expression of these genes in starved cells compared to counter non-starved cells.
Accordingly, the results make us believe that the cells are capable of gaining new characters during starvation, and the reduction of viable cell number in starved cultures incubated for an extended time did not decrease the mRNA levels of HSPs and CSC markers since the measurement burgeoned from the same starter mRNA quantities. The same was true for the non-starved cultures that incubated for an extended time (more than 24 h), the cultures mRNA levels of genes under investigation were higher during the extended time of incubation in comparison with cultures of 24 h incubation time. These cells will suffer from starvation as a result of gradual nutrient consumption in their cultures when incubation time expanded [8].
The ABCG2 is a transporter protein, as a member of the ATPbinding cassette transporters family, its play an important role in normal cells as efflux pump that transport byproducts, toxins, drugs, and other unwanted materials from the cytoplasm to the out of the cell. Consequently, this gene has an essential function in cellular physiology. Recently this gene was shown to play a pivotal role in cancer cell stemness [32]. In the situation present here, the extended time incubation of SKGT-4 cells with or without starvation would accumulate metabolic byproducts in the culture media that may enhance the induction of ABCG2 in the cells. That can be conceder as an environmental selective pressure, will eliminate cells which not capable of responding to it, leaving those with ABCG2 expression. The presence of this protein contributes to cell resistant to unfavorable stress render tumors capable or recurrent [33,34]. The aldehyde dehydrogenase (ALDH) is super-family of isoenzymes have an important function in protecting normal stem cells [35]. These enzymes contribute to stress resistant via the clearance of toxic byproducts of reactive oxygen species (ROS) from the internal of the cell [36]. Since the main manifestation of starvation and extended time of incubation in unchanged culture media in vitro was the accumulation of metabolic byproduct, the elevated level of this protein becomes deliverance. This explains the increment of this enzyme in cells under stressful environment makes them capable of remain in culture as a result of stress-resistant.
In conclusion, these results indicate that the stress cancer cells may encounter in their microenvironment would render them new capabilities such as that of stem cells in order to evade such stress, although they will definitely suffer from a severe loss in viability in their population either through apoptosis or necrosis.
Author Contributions
ATT designed experiments, ATT and NAM carried out the experiments, ATT, NAM, and ZS performed data analysis and results in interpretation, ATT, NAM, and ZS performed statistical analysis, ATT and NAM wrote the paper. All authors read and approved the final manuscript.
Acknowledgment
The authors would like to thank Miss Aimen Ali for her help in the maintenance and cultivation of the cell line. We thank Miss Rasha A. Hussein, Amina A. Yousif, and Teeba H. Jafer for their technical support. We thank Dr. Ahmed Al-Shimmary for supporting this research all the time and supplementing the cell line.
Data Availability Statement
The (histograms, gene names, images) data used to support the findings of this study are included in the article and can be provided to researchers at any inquiry.
References
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Nat Acad Sci 2003; 100: 3983-3988.
- Dang HT, Budhu A, Wang XW. The origin of cancer stem cells. J Hepatol 2014; 60: 1304-1305.
- Blazek ER, Foutch JL, Maki G. Daoy medulloblastoma cells that express CD133 are radioresistant relative to CD133-cells, and the CD133+ sector is enlarged by hypoxia. Int J Rad Oncol Biol Phys 2007; 67: 1-5.
- Mimeault M, Batra SK. New advances on critical implications of tumor-and metastasis-initiating cells in cancer progression, treatment resistance and disease recurrence. Histol Histopathol 2010; 25: 1057.
- Mimeault M, Batra SK. Complex oncogenic signaling networks regulate brain tumor-initiating cells and their progenies: pivotal roles of wild-type EGFR, EGFRvIII mutant and hedgehog cascades and novel multitargeted therapies. Brain Pathol 2011; 21: 479-500.
- Mimeault M, Batra SK. Novel biomarkers and therapeutic targets for optimizing the therapeutic management of melanomas. World J Clin Oncol 2012; 3: 32.
- Mimeault M, Batra SK. Frequent deregulations in the hedgehog signaling network and cross-talks with the epidermal growth factor receptor pathway involved in cancer progression and targeted therapies. Pharmacol Rev 2010; 62: 497-524.
- Tavaluc RT, Hart LS, Dicker DT, El-Deiry WS. Effects of low confluency, serum starvation and hypoxia on the side population of cancer cell lines. Cell Cycle 2007; 6: 2554-2562.
- Al-Saeedi ATT. Heat stress elevates cell population that express cancer stem cell markers in different types of cancer cell line. Mol Ther 2016; 24: 268.
- Plaks V, Kong N, Werb Z. The cancer stem cell niche: how essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015; 16: 225-238.
- Heddleston JM, Li Z, Lathia JD, Bao S, Hjelmeland AB, Rich JN. Hypoxia inducible factors in cancer stem cells. Br J Cancer 2010; 102: 789-795.
- De Miguel M, Alcaina Y, Sainz de la Maza D, Lopez-Iglesias P. Cell metabolism under microenvironmental low oxygen tension levels in stemness, proliferation and pluripotency. Curr Mol Med 2015; 15: 343-359.
- Chatterjee S, Burns TF. Targeting heat shock proteins in cancer: a promising therapeutic approach. Int J Mol Sci 2017; 18: 1978.
- Sevin M, Girodon F, Garrido C, de Thonel A. HSP90 and HSP70: implication in inflammation processes and therapeutic approaches for myeloproliferative neoplasms. Mediat Inflamm 2015; 2015.
- Roerecke M, Shield KD, Higuchi S, Yoshimura A, Larsen E, Rehm MX. Estimates of alcohol-related oesophageal cancer burden in Japan: systematic review and meta-analyses. Bull WHO 2015; 93: 329-338.
- Qian X, Tan C, Wang F, Yang B, Ge Y, Guan Z. Esophageal cancer stem cells and implications for future therapeutics. Oncotargets Ther 2016; 9: 2247.
- Wang D, Plukker JTM, Coppes R. Cancer stem cells with increased metastatic potential as a therapeutic target for esophageal cancer. Sem Cancer Biol 2017.
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res 2002; 12: 996-1006.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402-408.
- Weber R, Bertoni APS, Bessestil LW, Brasil BMdAA, Furlanetto TW. Validation of reference genes for normalization gene expression in reverse transcription quantitative PCR in human normal thyroid and goiter tissue. Biomed Research Int 2014; 2014.
- Strober W. Trypan blue exclusion test of cell viability. Curr Protocols Immunol 1997; 21.
- Thet MM, Yokote Y, Chu Q, Wang QT, Kishino K, Tamura N, Shimada J, Amano O, Kanda Y, Kunii S, Takekawa F, Hatano H, Shimada C, Hasegawa H, Sakagami H. Non-apoptotic cell death induced by nutritional starvation in J774.1 mouse macrophage-like cell line. Anticancer Res 2009; 29: 5083-5088.
- Sakagami H, Kishino K, Amano O, Kanda Y, Kunii S, Yokote Y. Cell death induced by nutritional starvation in mouse macrophage-like RAW264. 7 cells. Anticancer Res 2009; 29: 343-347.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646-674.
- La Porta CA. Thoughts about cancer stem cells in solid tumors. World J Stem Cells 2012; 4: 17-20.
- Tirino V, Desiderio V, Paino F, De Rosa A, Papaccio F, La Noce M, Laino L, De Francesco F, Papaccio G. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB J 2013; 27: 13-24.
- Najumudeen AK, Jaiswal A, Lectez B, Oetken-Lindholm C, Guzmán C, Siljamaki E, Posada IM, Lacey E, Aittokallio T, Abankwa D. Cancer stem cell drugs target K-ras signaling in a stemness context. Oncogene 2016; 35: 5248-5262.
- Ross RA, Walton JD, Han D, Guo H-F, Cheung N-KV. A distinct gene expression signature characterizes human neuroblastoma cancer stem cells. Stem Cell Res 2015; 15: 419-426.
- Tang DG. Understanding cancer stem cell heterogeneity and plasticity. Cell Res 2012; 22: 457-472.
- Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature 2013; 501: 328-337.
- Kim Y. The effects of nutrient depleted microenvironments and delta-like 1 homologue (DLK1) on apoptosis in neuroblastoma. Nutr Res Pract 2010; 4: 455-461.
- Wee B, Pietras A, Ozawa T, Bazzoli E, Podlaha O, Antczak C. ABCG2 regulates self-renewal and stem cell marker expression but not tumorigenicity or radiation resistance of glioma cells. Sci Rep 2016; 6: 25956.
- Zhang G, Wang Z, Luo W, Jiao H, Wu J, Jiang C. Expression of potential cancer stem cell marker ABCG2 is associated with malignant behaviors of hepatocellular carcinoma. Gastroenterol Res Pract 2013; 2013.
- Hu J, Li J, Yue X, Wang J, Liu J, Sun L, Kong D. Expression of the cancer stem cell markers ABCG2 and OCT-4 in right-sided colon cancer predicts recurrence and poor outcomes. Oncotarget 2017; 8: 28463-28470.
- Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev 2011; 7: 292-306.
- Levi BP, Yilmaz OH, Duester G, Morrison SJ. Aldehyde dehydrogenase 1a1 is dispensable for stem cell function in the mouse hematopoietic and nervous systems. Blood 2009; 113: 1670-1680.