ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2008) Volume 19, Issue 2
Simple method for speciation of clinically significant coagulase negative Staphylococci and its antibiotic sensitivity/resistant pat-tern in NICU of tertiary care centre.
1P.G. Department of Microbiology, C. S. M. Medical University, Lucknow, India
2Pediatrics Department, C. S. M. Medical University, Lucknow, India
3Department of Biochemistry, C. S. M. Medical University, Lucknow, India
- *Corresponding Author:
- R.K. Singh
Department of Biochemistry
CSMMU
Lucknow
India.
Mobile: 0091-9839829211
Fax: 0091-2257539
E-mail: singhrk23a@hotmail.com
Accepted date: April 21 2008
An attempt was made to speciate and antimicrobialy differentiate 150 clinically significant isolates of CoNS by practical scheme adapted from various references. Total 150 isolates were collected from NICU, Pediatrics Department, CSMMU, Lucknow from different sites and subjected to antimicrobial screening and biochemical characteriza-tion using conventional microbiological methods. Antibiotic sensitivity test was performed for all clinically significant isolates. 90% isolates were conveniently identified. S.epidermidis (40%), S.saprophyticus (14%), S.haemolyticus (12%), S.hominis (6%), S.lugdunensis (6%), S.schleiferi (2%), S.capitis (4%), S.warneri (6%), 15 isolates were not identified to the species level. Antibiotic susceptibility testing showed maximum resistance to ampicillin and penicillin with 80% and 38% strains showed the resistance to oxacillin. The increasing recognition of pathogen potential CoNS and emergence of drug resistance among them demonstrates the need to adopt simple laboratory procedure to identify and de-termine the prevalence and antibiotic sensitivity of CoNS. It will help in treating clinicians for first line treatment in hospital.
Keywords
Antibiotic susceptibility, clinical isolates, co-agulase negative staphylococci, identification, Kloos, Shleifer scheme for identification of bacteria.
Introduction
Co-agulase negative staphylococci have become common cause of nosocomial infections, particularly blood stream infection [1] and infections related to prosthesis [2]. They account for 9% of nosocomial infection [3]. The natural habitats of staphylococcus epidermidis and several other species of CoNS include skin and nares [4]; a clinical specimen could therefore, contain both contaminants and pathogens of these species. If contaminant is mistakenly identified as a pathogen i.e. source of isolate was the skin and not an infective process, the patient will probably receive unneeded antimicrobial therapy. It is now possible to identify the different species of staphylococcus by methods practical for diagnostic laboratories. Because of this more laboratories can access what potential clinical or epidemiological benefits this addition information might have.
The present study was aimed to identify the most preva-lent clinical isolates of CoNS by minimum number of tests necessary and sufficiently to discriminate between species. The tests, which were simple, inexpensive and easy to perform, were selected from the scheme of Kloos and Shleifer to identify CoNS species group (group ap-proach) or species [5,6]. Further one or two additional test was included to complete the strain identification wher-ever necessary. Antimicrobial susceptibility profiles of all isolates were done by Disk Diffusion Method.
Material and Methods
A total of 150 consecutive non-repeat clinically signifi-cant CoNS isolates were collected from January 2006 to December 2006 from Pediatrics Department and processed in the Department of Microbiology, Chhatrapati Shahuji Maharaj Medical University, Lucknow. Strains were isolated from blood sample, pus, urinary catheter tip and urine. The isolates were considered clinically signifi-cant when isolated in pure culture from infected site. The strains collected were initially identified by colony mor-phology, Gram staining, catalase, slide and tube coagu-lase (read after 4-24 hr) and anaerobic acid from manni-tol. Bacitracin (0.04U) susceptibility was done to exclude micrococcus and stomatococcus spp [5,7].
Identification
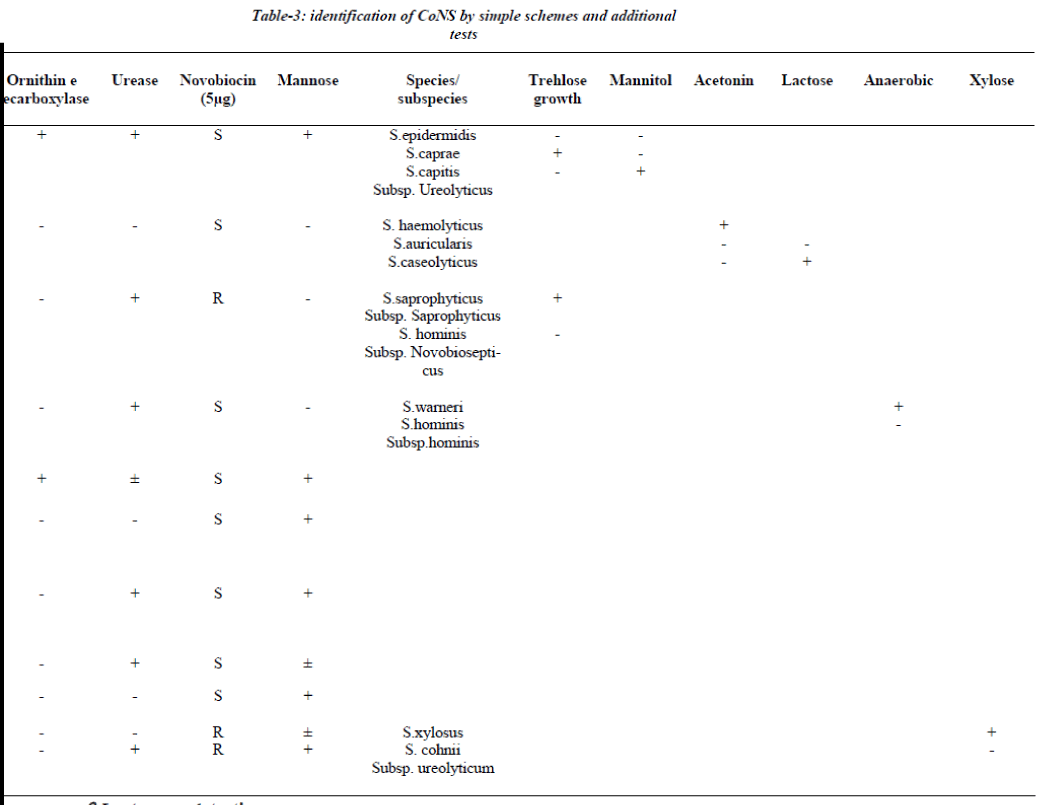
All the strains, which were either, slide or tube coagulase negative were further identified by a scheme [3,8,-12]. The identification scheme concentrated on spe-cies groups/ species commonly encountered in clinical practice the S. epidermidis group (i.e. S.epidermidis, S. capitis subspecies urolyticus and caprae), the S. haemolyticus group (S. haemolyticus, S. auricularis and S.caseolyticus), the S.saprophyticus group (S .saprophyti-cus subspecies saprophyticus and S.hominis subspecies novobiosepticus), the S.warneri group (S.warneri and S.hominis subspecies hominis), the S.cohnii group (S.xylosus and S. cohnii subspecies urolyticum), S. lug-dunensis , S.shleiferi subspecies schleiferi, S.capitis sub- species capitis, S.simulans and S.cohnii subspecies cohnii [6]. This scheme involves a two-step procedure (Table-1), first step aimed to identify species group and combined slide and tube coagulase with novobiosin resistance test for urease activity, ornithine decarboxylase & aerobic acid production from mannose. If identification requires additional tests, a maximum of two tests were selected from Table 1: trehalose and mannitol for the S. epidermidis group, acetoin production and lactose for S.haemolyticus group, trehalose for the S.saprophyticus group, anaerobic thioglycollate both for S. warneri group and xylose for S. cohnii group [6]. The entire tests were performed according to reference method [5].
Antimicrobial susceptibility test
The susceptibility of the isolates to antimicrobial agents, including oxacillin was determined by the disk diffusion method with Mueller-Hinton agar plates (Hi-Media, Mumbai) according to the guidelines NCCLS [13,14]. Disk contain following antibiotic at the specific absolute concentrations penicillin 10μg, ampicillin 10μg, chloram-phenicol 30μg, erythromycin 15μg, rifampicin 5μg, gen-tamycin 10μg, vancomycin 30μg, oxacillin 1μg, staphy-lococcus aureus ATCC 43300 was used as control.
β Lactamase detection
Acidometric determination of β Lactamase production was performed by using growth from around the oxacillin disk.
Result
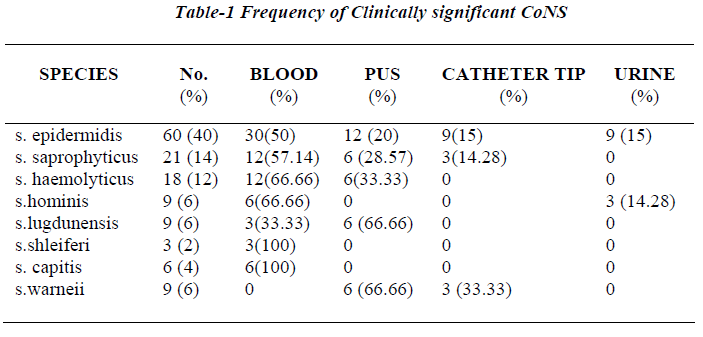
Seventy two out of 150 strains of CoNS (60%) were iso-lated from blood samples, 36 from pus samples, 15 from urinary catheter tip and 12 from the urine samples. All 150 strains were negative for blood clumping factor and tube coagulase negative. Table 3 shows the CoNS species group /species by the scheme [15].
The scheme could identify 90% of CoNS isolated from clinical samples incorporation of one or two additional test whenever needed. S. epidermidis was the most fre-quent isolate and was identified if ornithine decarboxylase was positive (60 isolates), while ornithine decarboxylase negative isolates required inclusion of trehlose and mannitol for speciation. 15 isolates were unidentified because of aberrant characteristics.
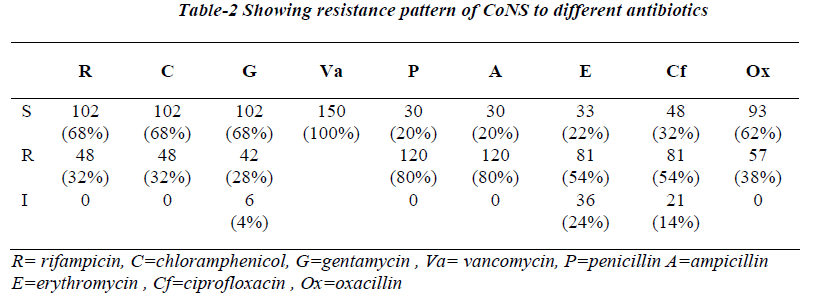
Antibiotic susceptibility testing showed maximum resis-tance to penicillin (80%) and ampicillin (80%) followed by erythromycin (54%), ciprofloxacin (54%), oxacillin (38%), rifampicin (32%), chloramphenicol (32%), genta-mycin (28%), no resistance to vancomycin was seen.
Resistance to oxacillin was highest among isolates of S.haemolyticus (76%) followed by S.epidermidis (33%). S.haemolyticus was most resistance species for both erythromycin (80%) and rifampicin (42%). S.epidermidis and S.hominis also showed a significant percentage of isoltes resistance to both of these agents. Resistance to gentamycin (76%) and ciprofloxacin (42%) was greatest for s.haemolyticus followed by S.saprophyticus. Resis-tance to chloramphenicol, ampicillin and penicillin was greatest for S.epidermidis.
Discussion
Many laboratories do not identify clinical isolates of CoNS to the species level as they are considered normal inhibitant of skin and nares capable of causing only op-portunistic infections [9]. Moreover, the conventional identification methods, though accurate, are cumbersome and employ a large battery of biochemical reactions, which often gives variable results and all the tests are generally not available in most of the routine diagnostic laboratories [16]. As CoNS is increasingly being impli-cated as significant nosocomial pathogen, several review-ers have emphasized the need for species identification, which is possible only by simple, easily adaptable, inex-pensive method [3,5,6,15,18,19]. The species identifica-tion is important in monitoring the reservoir and distribu-tion of CoNS involved in nosocomial infections and de-termining the etiological agent [5,15]. The present scheme conveniently identified the most frequently encountered clinical isolates in our hospital as S.epidermidis (40%), S.haemolyticus (12%), S. saprophyticus (14%), S. hominis (6%), S.lugdunensis (6%) upto species level by incorpora-tion of one or two additional tests. This scheme could directly identify even the newly described species S.schleiferi subsp. Schleiferi and S. lugdunensis using slide agglutination test for detection of clumping factor or fibrinogen affinity factor provided no slide is discarded before 10 seconds as S.lugdunensis sometimes gives de-layed results [5]. Consequently, the laboratories perform-ing only slide agglutination test without tube coagulase test may misidentify S.schleiferi subsp. Schleiferi and S. lugdunensis as S.aureus. This scheme could not differen-tiate S.capitis subsp. Ureolyticus, S.warneri, S.simulans as pyroglutamyle β napthylamide (PYR) hydrolysis test, a key component for their identification, being expensive was not included. Various workers from India have re-ported S.epidermidis and S.saprophyticus to be the most common isolate similar to our study [9,10,17,18]. Though a single report has mentioned isolation of S.cohnii which was not isolated in our study, however, the scheme described can identify this isolate [9].
This scheme identified 90% of CoNS upto the species level which is almost similar to earlier report [9,10,17-19]. The most obvious finding was observed in smaller numbers by others [21]. Out of all species S. haemolyticus has the most antibiotic resistant profile. Ap-proximately 76% of our isolates of S. haemolyticus showed resistance to oxacillin (76%), gentamycin (76%), erythromycin (80%). Although, S.epidermidis can also show significant multiple resistance patterns. It is there-fore recommended to assess the importance of CoNS speciate whatever level possible and perform the antibi-otic susceptibility testing before any typing procedure for epidemiological studies are being undertaken. This simple, inexpensive methodology will prove useful in routine microbiology laboratory for the presumptive identification of CoNS.
Acknowledgement
The authors are thankful to Mr. R. K. Tiwari, Laboratory Technician, Bacteriology Laboratory, Department of Mi-crobiology, C.S.M.M.U., Lucknow for technical assistance.
References
- Pfaller MA, Jones RN, Doern GV, Sader HS, Kugler KL, Beach ML, the SENTRY Participants Group. Sur- vey of bloodstream infections attributable to gram- positive cocci: frequency of occurrence and antimicro- bial susceptibility of isolates collected in 1997 in United States, Canada and Latin America from the SENTRY Antimicrobial Surveillance Program. Diagn. Microbiol Infect Dis 1999. 33: 283-297.
- Rupp ME, GL Archer. Coagulse-negative staphyloco- cci: pathogens associated with medical progress. Clin Infect Dis 1994 19: 231-245.
- Kloos WE, Bannerman TL. Update on clinical significance of coagulase negative staphylococci. Clin Micro- bial Rev 1994; 7:117-140.
- Kloos WE. Natural populations of the genus Staphylococcus. Annu. Rev. Microbiol 1980; 34: 559-592.
- Bannerman TL. Staphylococcus, Micrococcus and other catalase positive cocci that grow aerobically, Chapter 28. In: Manual of Clinical Microbiology, 8th ed. Murray PR, Baron EJ, Jorgensen JH, Pfaller MA, Yolken RH, editors. ASM press: Washington DC; 2003 p.384
- De paulis AN, Predari SC, Chazarreta CD, Santoiani JE. Five test simple scheme for the species level identi- fication of clinically significant coagulase negative staphylococci. J.Clin Microbiol 2003; 41: 1219-1224.
- Baird D. Staphylococcus; cluster forming Gram-posi- tive cocci, chapter 11. In: Mackie And Mc Cartney Pra- ctical Medical Microbiology, 14th ed. Collee JG, Fraser AG, Marimom BP, Simmons A, editors. Churchill Liv- ingstone: New York: 1996. p. 245.
- Ieven M, Verhoeven J, Pattyn SR, Goossens H. Rapid and economical method for species identification of clinically significant Cogulase negative Staphylococci. J. Clin.Microbiol 1995; 33: 1060-1063.
- Shrikhande S, Thakkar YS, Pathak AA, Saoji AM. Species distribution of clinical isolates of Staphylo- cocci. Indian J Med Microbiol 1996; 39: 207-210.
- Phatak J, Udgaonkar U, Kulkarni RD, Pawar SG. Study of cogulase negative staphylococci and their incidence in human infections. Indian J Med Microbiol 1994; 12: 90-95.
- Hebert GA, Crowder CG, Hancock GA, Jarvis WR, Thornsberry C. Charecterstics of Coagulase negative Staphylococci that help differentiate these species and other members of family Micrococcaceae. Clin Micro- biol 1998; 26:1939-1949.
- Goyal R, Singh NP, Kumar A, Kaur I, Singh M, Sunita N, Mathur M. Simple and economical method for spe- ciation and resistotyping of clinically significant coagu- lase negative staphylococci, Indian J Med Microbiol 2006, 24: 201-2044.
- National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk suscepti- bility test, 7th ed. NCCLS document M2-A7. National Committee for Clinical Laboratory Standards, 2000. Wayne, Pa.
- National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing: twelfth informational supplement. NCCLS document M100-S12. National Committee for Clinical Laboratory Standards, 2002 Wayne, Pa.
- Archer GL, climo MW. Antimicrobial susceptibility of coagulase negative staphylococci. Antimicrobial. Agent chemother 1994; 38: 2231-7.
- National Committee for Clinical Laboratory Standards (NCCLS). Performance standards for antimicrobial disk susceptibility tests: approved standards. NCCLS document. NCCLS: Wayne, Pa.2000.
- Mohan V, jindal N, Agarwal P. Species distribution and antibiotic sensitivity pattern of co- agulase negative staphylococci isolated from various clinical specimens . Indian J. Med. Microbiol. 2002; 20: 45-46.
- Saini S, Kaur H, Sabharwal U, Malik AK. Coagulase negative staphylococci in the urinary tract. Indian J Med Res 1983; 78: 26-28.
- Goel MM, Singh AV, Mathur SK, Singh M, Singhal S., Chaturvedi UC. Resistant coagulase negative staphylo- cocci from clinical samples. Indian J Med Res 1991; 93: 350-352.
- Price SB, Flournoy DJ. Ccomparison of antimicrobial susceptibility patterns among coagulase negative staph- ylococci. Antimicrob. Agent chemother 1982; 21: 436-440.