ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2019) Volume 30, Issue 4
Mining hub genes associated with late stage adrenocortical carcinoma
Di Yu1,2, Chen Dongshan1,2, Yu Wei3, Yan Lei1*
1Department of Urinary Surgery, Qilu Hospital, Jinan, Shandong, P.R. China
2Department of Shandong Cardiovascular Research Center, Shandong University, Jinan, Shandong, P.R. China
3Department of Urinary Surgery, Lanzhou University, P.R. China
- *Corresponding Author:
- Yanlei
Department of Urinary Surgery
Qilu Hospital, Jinan Shandong P.R. China.
Accepted Date: May 20, 2019
DOI: 10.35841/biomedicalresearch.30-19-247
Visit for more related articles at Biomedical ResearchBackground: Adrenocortical carcinoma (ACC) was in poor prognosis especially the late stages. Our research tried to discover potential hub genes associated with the disease. Methods: The GSE90713 including 5 normal and 58 ACC samples were downloaded from the GEO datasets. A total of 106 up- and 299 down-regulated DEGs were identified by BrB-ArrayTools. Then their gene ontology functions and KEGG pathways were enriched by the DAVID. The protein-protein interaction network was provided by STRING. Then, Cytohubba was used to pick out the hub genes. The Webgestalt was used to predict transcription factors and microRNAs. Finally, TCGA’s data was utilized to validate our results. Results: The up-regulated DEGs were mainly involved in the processes of cell division and cell cycle. The down regulated DEGs were mainly involved in the processes of response to hormone and extracellular exosome. CDK1, RFC4, KIF11, TOP2A, CCNB1, MAD2L1, AURKA, NCAPG, CDKN3, TRIP13, were identified hub genes and they were closely related to the process of the mitotic cell cycle. The high expression of these genes indicated poor prognosis of the diseases (P<0.01) by Kaplan Meier test. After examination by the genome data of ACC patients in TCGA datasets, they were suggested not only associated with the metastasis, but also with the tumor staging. However, except CCNB1, KIF11, MAD2L1, and TRIP13, others didn’t perform an obvious connection with local lymph nodes invasion. Conclusion: CCNB1, KIF11, MAD2L1, and TRIP13 were closely associated with TMN stage and prognosis. They may serve as therapeutic targets for the adrenal carcinoma.
Keywords
Adrenocortical cancer, Cytohubba, Hub genes, Late stage, Prognosis.
Abbreviations
ACC: Adrenal Cortical Carcinoma; DEGs: Differential Expressed Genes; STRING: Search Tool for the Retrieval of Interacting Genes; TCGA: The Cancer Genome Atlas; EPC: Degree-Edge Percolated Component; MNC: Maximum Neighborhood Component; DMNC: Density of Maximum Neighborhood Component; MCC: Maximal Clique Centrality; GO: Gene Ontology; MF: Molecular Function; BP: Biological Process; TF: Transcriptional Factors; KIF11: Kinesin Family Member 11; TOP2A: Topoisomerase (DNA) II alpha; CCNB1: Cyclin B1; MAD2L1, MAD2 Mitotic Arrest Deficient-Like 1; CDKN3: Cyclin-Dependent Kinase Inhibitor 3; CDK1: Cyclin-Dependent Kinase 1; AURKA: Aurora Kinase A; NCAPG: Non-SMC Condensin I Complex Subunit G; TRIP13: Thyroid Hormone Receptor Interactor 13; RFC4: Replication Factor C 4; PCNA: Proliferating Cell Nuclear Antigen; MPF: Maturation-Promoting Factor.
Introduction
Adrenocortical carcinoma was a rare disease in which malignant cells derived from the outer layer of the adrenal gland [1]. Although very rare-only 5 to 10 percent of all adrenal tumors were malignant-they were often aggressive and can cause many symptoms and discomforts, such as pain in the abdomen [2]. Imaging studies and tests examining the blood and urine were often used to detect and diagnose adrenocortical carcinoma [3]. The stage including the size of the tumor, the local lymph node invasion and whether it is in the adrenal gland only or has spread to other places in the body, seriously affected the patient’s prognosis. Most adrenocortical tumors were functioning and having certain genetic conditions may increase the risk of adrenocortical carcinoma. The hormones made by functioning tumors may cause certain signs or symptoms of the disease [4,5]. The prognosis and treatment options depended on the tumor’s stage, the possibility to be completely removed in surgery, as well as the patient's general health and so on. When the tumor had metastasized to other organs, the patients may suffer a poor living quality [6]. However, with the developing of computer science, the bioinformatics was widely used in the medical field. The accuracy therapy for the cancers was also thriving in recent years [7], which was also suitable for the late stage ACCs when surgery effect was limit. However, hub genes identified by bio-informatic methodologies in ACCs were still lack of literature reports, especially about the TMN staging. In our article, we tried to use BrB-ArrayTools to find the DEGs and Cytohubba to discover the key genes. Then, the data in the TCGA was used to validate our results. What’s more, their miroRNAs and TFs were predicted by the Webgestalt visualized by Cytoscape.
Materials and Methods
Data description
The gene chip of GSE90713 was obtained from the GEO database [8]. Its platform was Affymetrix Human Gene Expression Array GPL15207. It consisted of 58 adrenocortical carcinoma samples and 5 normal adrenal samples in total. The data has been preprocessed and its distribution in GEO2R was uniform and suitable for analyzing.
DEGs identification
BRB-ArrayTools was a convenient tool for bioinformatic analysis [9,10]. We imported the expression matrix into BRB-Arraytools, exclude a gene if minimum fold change is less than 20% of an expression data value or have at least a 1.5-fold change in either direction from the gene median value or percent missing exceeds 50%. Finally, the significance threshold of univariate tests is set of 0.001 by random variance model and the groups of the normal tissue and carcinoma tissue were compared to identify the DEGs [11].
Gene ontology function annotation and pathways analysis of the DEGs
DAVID was a free online bioinformatics resource developed by the Laboratory of Immunopathogenesis and Bioinformatics [12]. All tools in the DAVID bioinformatics resources provided a functional interpretation of large lists of genes derived from genomic studies, including gene ontology and pathways analysis. Similarity, BiNGO, the plugin of Cytoscape, and Panther also provide the service of the gene ontology analysis [13,14].
PPI network construction and hub genes identification
The interaction network was obtained by uploading the DEGs to the Search Tool for the Retrieval of Interacting Genes database [15]. Then the interaction file and annotation table were input into the Cytoscape software version 3.6.0, the PPI network can be professionally visualized. The hub genes are identified by Cytohubba, which can explore important nodes and fragile motifs in an interactome network by several topological algorithms including degree-edge percolated component (EPC), maximum neighborhood component (MNC), density of maximum neighborhood component (DMNC), maximal clique centrality (MCC) and so on. In our research, the top 10 genes were identified by their MCC values [16,17].
TFs and microRNA network construction
The transcriptional factors (TFs) and microRNAs of targeted DEGs were predicted by the Webgestalt. WebGestalt supports three well-established and complementary methods for enrichment analysis, including Over-Representation Analysis (ORA), Gene Set Enrichment Analysis (GSEA), and Network Topology-based Analysis (NTA). The up and down-regulated DEGs were separately inputted into the website and their sharing TFs and microRNAs were inferred by from comparative genomic analysis and made available through MSigDB [18,19].
Hub genes function annotation
Given a set of genes to GenClip2.0, for example from high-throughput experiments, it can be helpful to know which biological functions and molecular networks may be involved, or whether genes from a given list are related to certain topics, such as various biological and pathological processes. The hub genes were then annotated by the tools [20].
Survival analysis of the obtained hub genes
The GEPIA, a portal using Genomics data from GTEx and The Cancer Genome Atlas (TCGA) project, was used to do Kaplan Meier test of these genes based on low and high expression. The hub genes acquired by Cytohubba were input into the website and their Kaplan Meier plots of overall survival and disease-free survival were obtained by the website tools [21,22].
Validation of the obtained hub genes in TNM staging of ACCs
Genomic data from The Cancer Genome Atlas (TCGA) project had been used to validate our analysis results. As mentioned above, the ACC patient’s data was downloaded from the website. We compared these genes’ expression by Wilcoxon test which grouped in M0 (no metastasis) and M1 (metastasized to other organs and distal lymph nodes) status [21]. Then, these genes’ association with the N0 (no local lymph nodes invasion) and N1 (local lymph nodes invasion) of ACC’s patient were examined. Finally, the Kruskal−Wallis test was done to validate the genes’ expression to the tumor stage.
Results
DEGs identification
A total of 405 DEGs of ACC samples were identified by comparing to the normal samples, consisting of 106 up- and 299 down DEGs.
Gene ontology and pathway enrichment analysis results
The gene ontology (GO) categories were classified into three groups: biological process, cellular component and molecular function. The GO terms and KEGG pathways were ranked by p-value and the top 5 were selected. The results of the upregulated DEGs were shown in Table 1 and the results of the down regulated DEGs were shown in Table 2. The upregulated DEGs were mainly involved in the processes of cell division, G1/S transition of the mitotic cell cycle, anaphasepromoting complex-dependent catabolic process, mitotic spindle organization, and DNA repair. The down regulated DEGs were mainly involved in the processes of response to estrogen and the peptide’s hormone, outflow tract septum morphogenesis, regulation of cell growth and erythrocyte homeostasis. Most of the up-regulated DEGs were involved in the cell cycle, Oocyte meiosis, p53 signaling, DNA replication, and progesterone-mediated oocyte maturation pathways. Most of the down regulated DEGs were involved in fatty acid degradation metabolic, metabolic, carbon metabolism, seleno-compound metabolism, and mineral absorption pathways.
| Term | p-value | Term | p-value |
|---|---|---|---|
| BP | MF | ||
| GO: 0051301~cell division | 1.38E-09 | GO: 0005524~ATP binding | 3.33E-05 |
| GO: 0000082~G1/S transition of mitotic cell cycle | 6.49E-07 | GO: 0035173~histone kinase activity | 1.46E-04 |
| GO: 0031145~anaphase-promoting complex-dependent catabolic process | 2.36E-06 | GO: 0042803~protein homodimerization activity | 2.53E-04 |
| GO: 0007052~mitotic pindle organization | 1.31E-05 | GO: 0005515~protein binding | 0.002331 |
| GO: 0006281~DNA repair | 1.91E-05 | GO: 0003677~DNA binding | 0.003263 |
| CC | KEGG pathway | ||
| GO: 0005654~nucleoplasm | 2.19E-10 | hsa04110: cell cycle | 6.28E-07 |
| GO: 0005634~nucleus | 5.47E-09 | hsa04114: Oocyte meiosis | 5.04E-05 |
| GO: 0005876~spindle microtubule | 6.07E-08 | hsa04115: p53 signaling pathway | 6.97E-04 |
| GO: 0005829~cytosol | 8.27E-05 | hsa03030: DNA replication | 0.001314 |
| GO: 0005737~cytoplasm | 9.38E-04 | hsa04914: Progesterone-mediated oocyte maturation | 0.015727 |
BP: Biological Process; CC: Cellular Components; MF: Molecular Functions.
Table 1. The top 5 enriched gene ontology functions and KEGG pathways of the up-regulated DEGs, *The terms are ranked by p-value.
| Term | P-Value | Term | p-value |
|---|---|---|---|
| BP | MF | ||
| GO: 0043627~response to estrogen | 0.00742 | GO: 0009055~electron carrier activity | 0.001068 |
| GO: 0043434~response to peptide hormone | 0.00275 | GO: 0050660~flavin adenine dinucleotide binding | 0.001381 |
| GO: 0003148~outflow tract septum morphogenesis | 0.0034 | GO: 0003779~actin binding | 0.003269 |
| GO: 0001558~regulation of cell growth | 0.00431 | GO: 0031995~insulin-like growth factor II binding | 0.004377 |
| GO: 0034101~erythrocyte homeostasis | 0.00599 | GO: 0001968~fibronectin binding | 0.004379 |
| CC | KEGG pathway | ||
| GO: 0070062~extracellular exosome | 1.30E-07 | hsa00071: Fatty acid degradation | 0.003194 |
| GO: 0005789~endoplasmic reticulum membrane | 3.81E-04 | hsa01100: Metabolic pathways | 0.004151 |
| GO: 0005739~mitochondrion | 5.53E-04 | hsa01200: Carbon metabolism | 0.024917 |
| GO: 0005615~extracellular space | 0.001376 | hsa00450: Selenocompound metabolism | 0.02515 |
| GO: 0005576~extracellular region | 0.003318 | hsa04978: Mineral absorption | 0.026343 |
BP: Biological Process; CC: Cellular components; MF: Molecular Functions.
Table 2. The top 5 enriched gene ontology functions and KEGG pathways of the down-regulated DEGs, *The terms are ranked by p-value.
PPI network construction and analysis
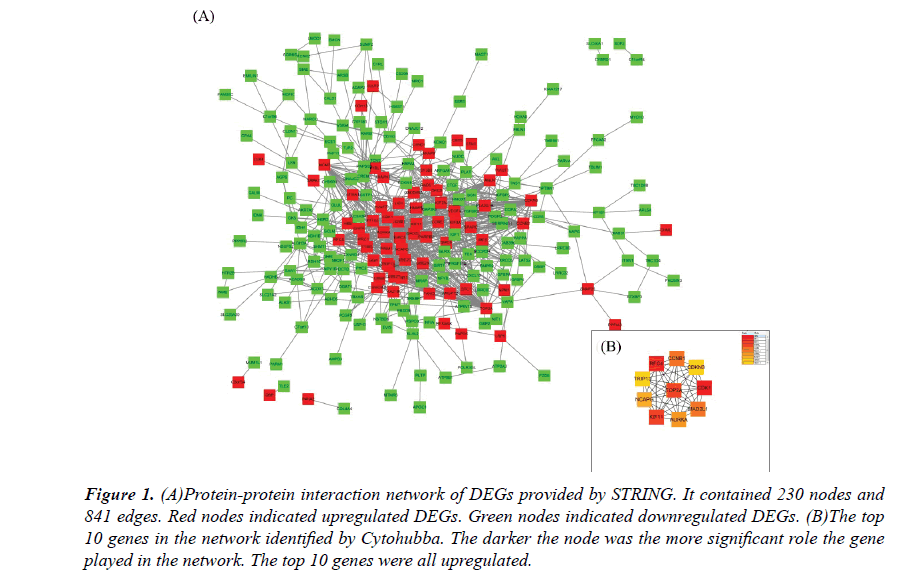
The PPI network constructed by the Cytoscape, as shown in Figure 1A. The PPI network included 230 nodes and 841 interactions, involving 70 up and 160 downregulated genes. The hub genes of the network were identified by Cytohubba, a plugin in the Cytoscape. The results were shown in Figure 1B. As ordered by MCC values, the top 10 hub genes were s.
Figure 1: (A)Protein-protein interaction network of DEGs provided by STRING. It contained 230 nodes and 841 edges. Red nodes indicated upregulated DEGs. Green nodes indicated downregulated DEGs. (B)The top 10 genes in the network identified by Cytohubba. The darker the node was the more significant role the gene played in the network. The top 10 genes were all upregulated.
TFs and microRNA network construction
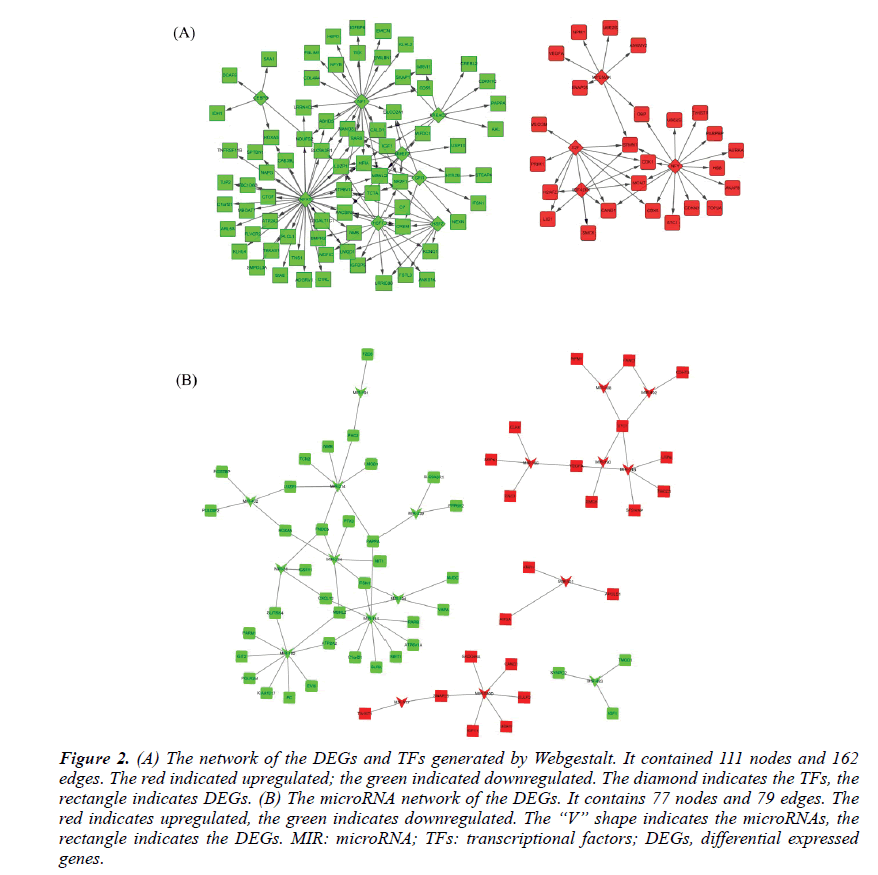
The TFs and microRNAs were inferred by Webgestalt tools with the overrepresentation enrichment analysis method. The FDR threshold was set of 0.01. After screening and filtering, a total of 12TFs and 18 microRNAs were suggested. The up-regulated TFs were E2F, E2F4DP1, MYCMAX, NFY. The down-regulated TFs were HSF1, NFAT, CEBPB, HSF2, MMEF2, NF1, FREAC2, and GFI1. Their network with DEGs was shown in Figure 2A. The upregulated microRNAs were hsa-mir-492, hsa-mir-488, hsa-mir-34b, hsa-mir-520d, hsa-mir-517, hsa-mir-501, hsa-mir-190, hsa-mir-186. The downregulated microRNAs were hsa-mir-31, hsa-mir-224, hsa-mir-432, hsa-mir-141, hsa-mir-191, hsa-mir-182, hsamir- 483, hsa-mir-214, hsa-mir-194 and hsa-mir-339. Their network with the DEGs was shown in Figure 2B.
Figure 2: (A) The network of the DEGs and TFs generated by Webgestalt. It contained 111 nodes and 162 edges. The red indicated upregulated; the green indicated downregulated. The diamond indicates the TFs, the rectangle indicates DEGs. (B) The microRNA network of the DEGs. It contains 77 nodes and 79 edges. The red indicates upregulated, the green indicates downregulated. The “V” shape indicates the microRNAs, the rectangle indicates the DEGs. MIR: microRNA; TFs: transcriptional factors; DEGs, differential expressed genes.
Hub genes function annotation
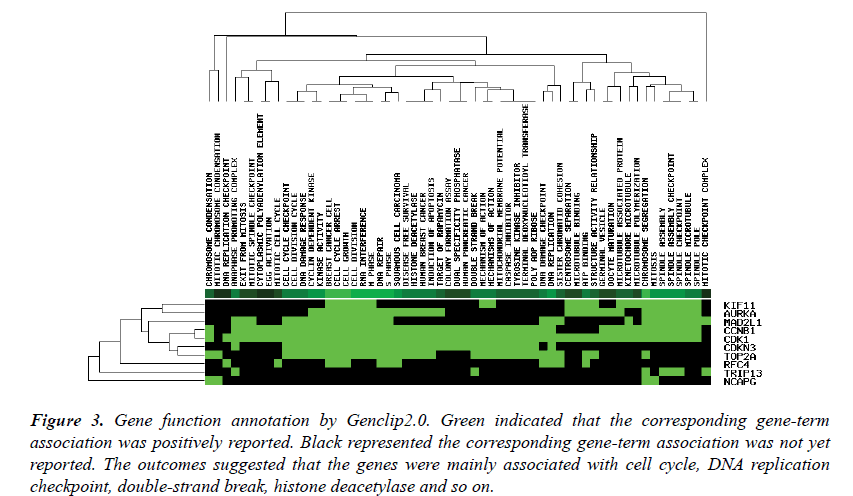
The top ten genes: CDK1, RFC4, KIF11, TOP2A, CCNB1, MAD2L1, AURKA, NCAPG, CDKN3, TRIP13, were input into the Genclip2.0. They were clustered with literature profile. The result was shown in Figure 3. They were associated with the process of the mitotic cell cycle, terminal deoxynucleotidyl transferase, apoptosis, caspase inhibitor and so on.
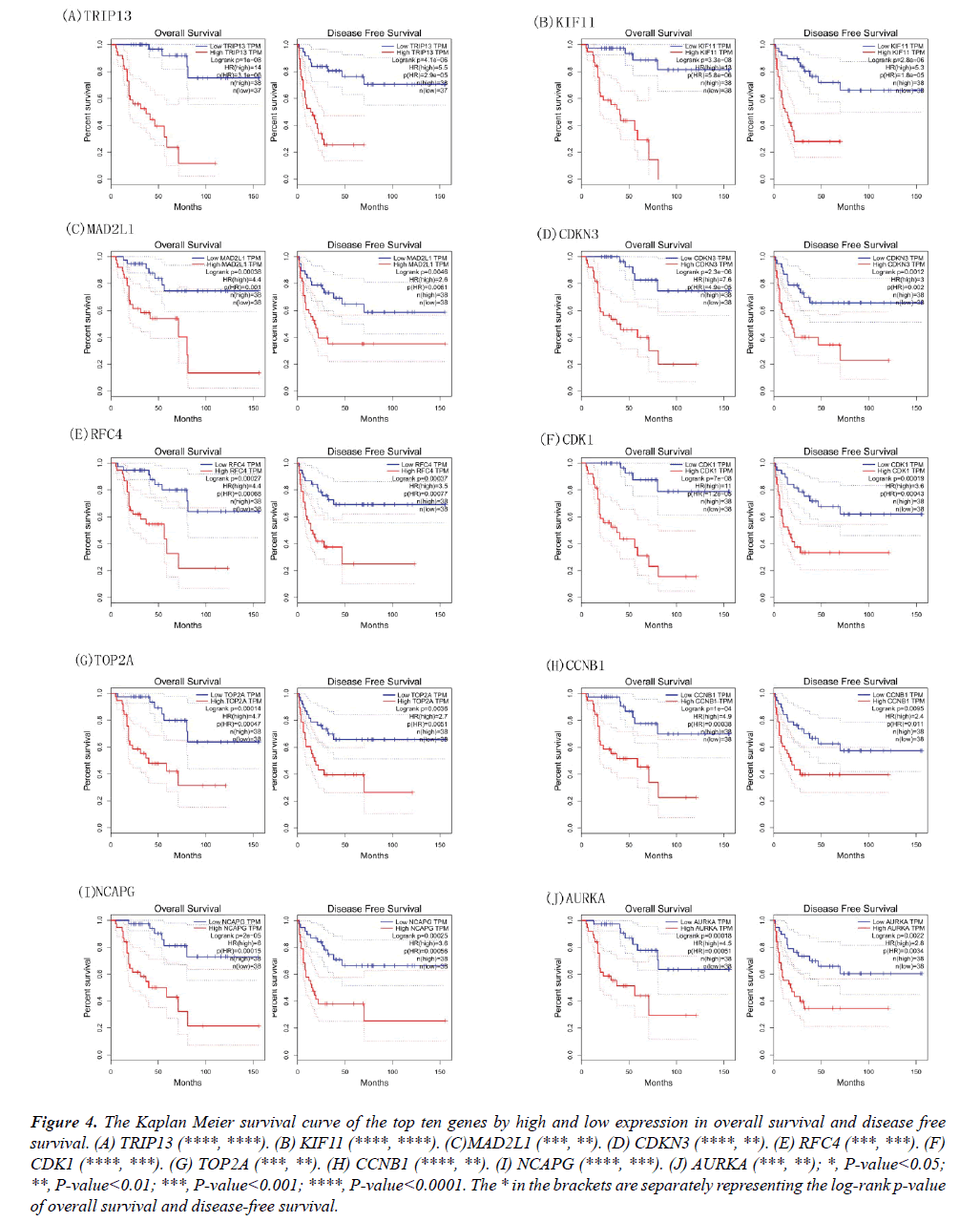
Figure 3: The Kaplan Meier survival curve of the top ten genes by high and low expression in overall survival and disease free survival. (A) TRIP13 (****, ****). (B) KIF11 (****, ****). (C)MAD2L1 (***, **). (D) CDKN3 (****, **). (E) RFC4 (***, ***). (F) CDK1 (****, ***). (G) TOP2A (***, **). (H) CCNB1 (****, **). (I) NCAPG (****, ***). (J) AURKA (***, **); *, P-value<0.05; **, P-value<0.01; ***, P-value<0.001; ****, P-value<0.0001. The * in the brackets are separately representing the log-rank p-value of overall survival and disease-free survival.
Survival analysis of the obtained hub genes
The Kaplan Meier curves of these genes were also obtained based on the high and low/medium expression; the results of overall survival and disease-free survival were shown in Figure 4. The high expression of CDK1, RFC4, KIF11, TOP2A, CCNB1, MAD2L1, AURKA, NCAPG, CDKN3, TRIP13 indicated low days on ACC patient survival. The genes of MAD2L1, CDK1, RFC4, KIF11, TOP2A, CCNB1, AURKA, NCAPG, CDKN3, TRIP13 may serve as prognostic biomarkers for the disease.
Figure 4: The Kaplan Meier survival curve of the top ten genes by high and low expression in overall survival and disease free survival. (A) TRIP13 (****, ****). (B) KIF11 (****, ****). (C)MAD2L1 (***, **). (D) CDKN3 (****, **). (E) RFC4 (***, ***). (F) CDK1 (****, ***). (G) TOP2A (***, **). (H) CCNB1 (****, **). (I) NCAPG (****, ***). (J) AURKA (***, **); *, P-value<0.05; **, P-value<0.01; ***, P-value<0.001; ****, P-value<0.0001. The * in the brackets are separately representing the log-rank p-value of overall survival and disease-free survival.
Validation of the obtained hub genes in the TNM staging of ACCs.
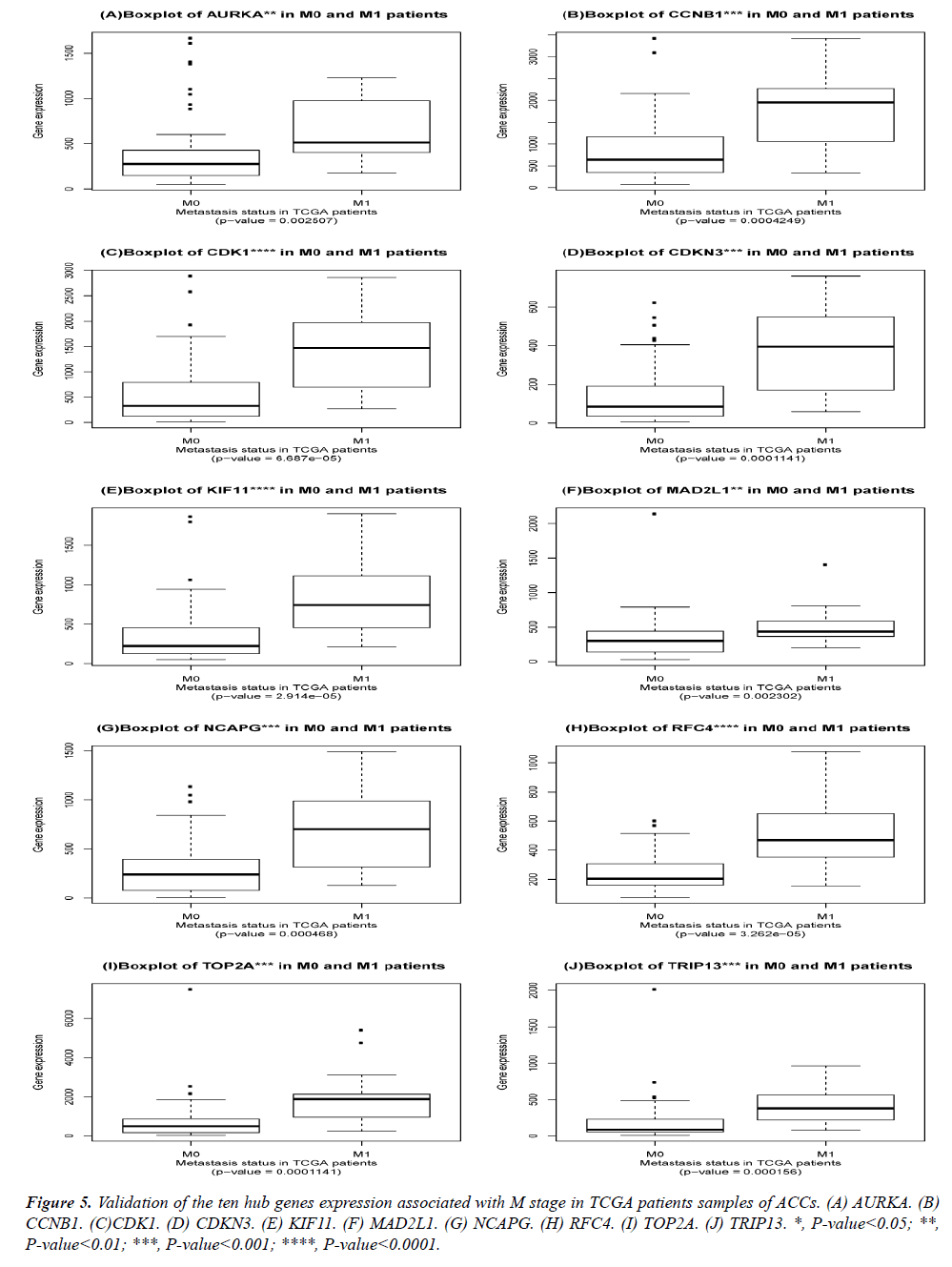
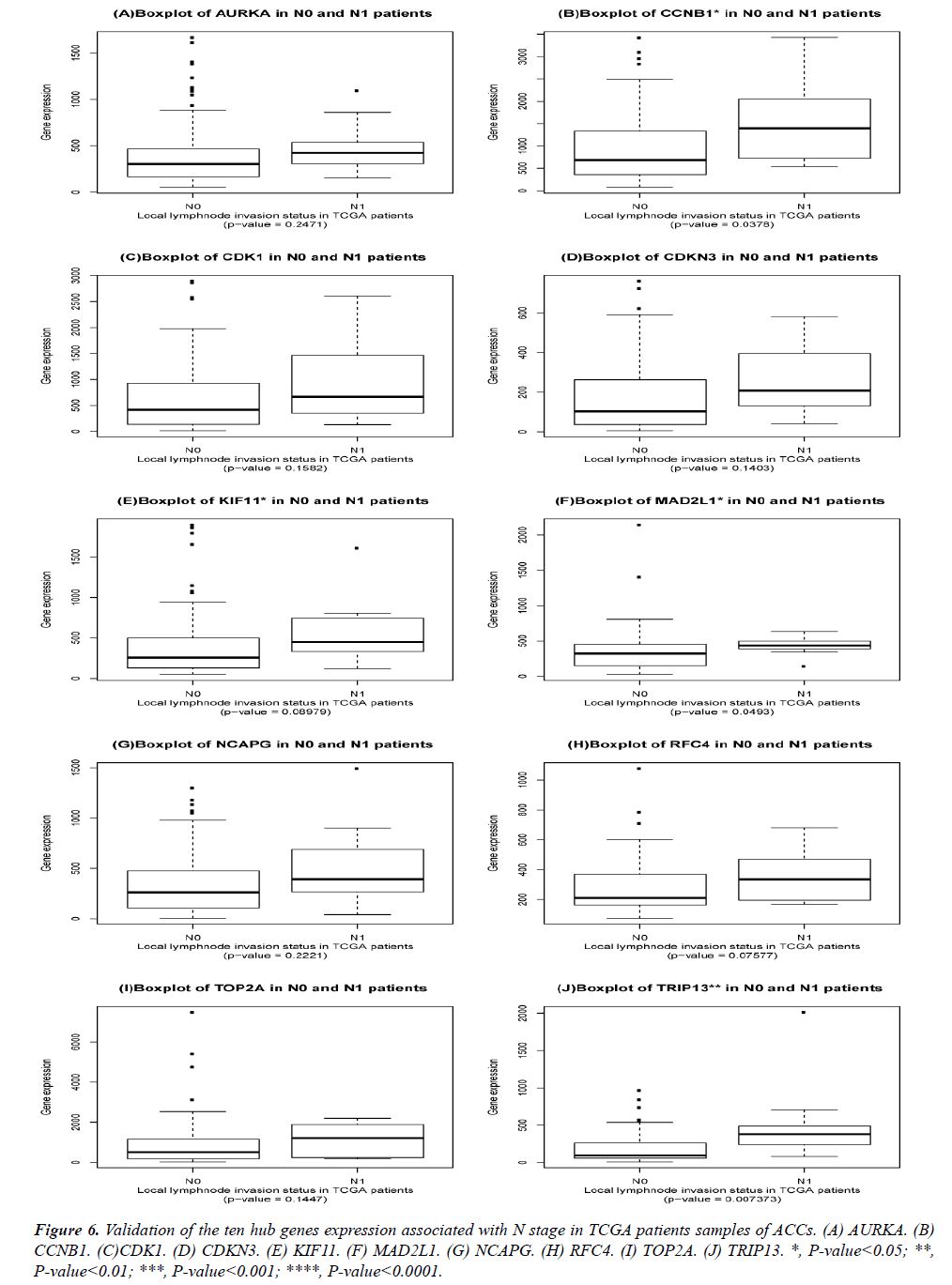
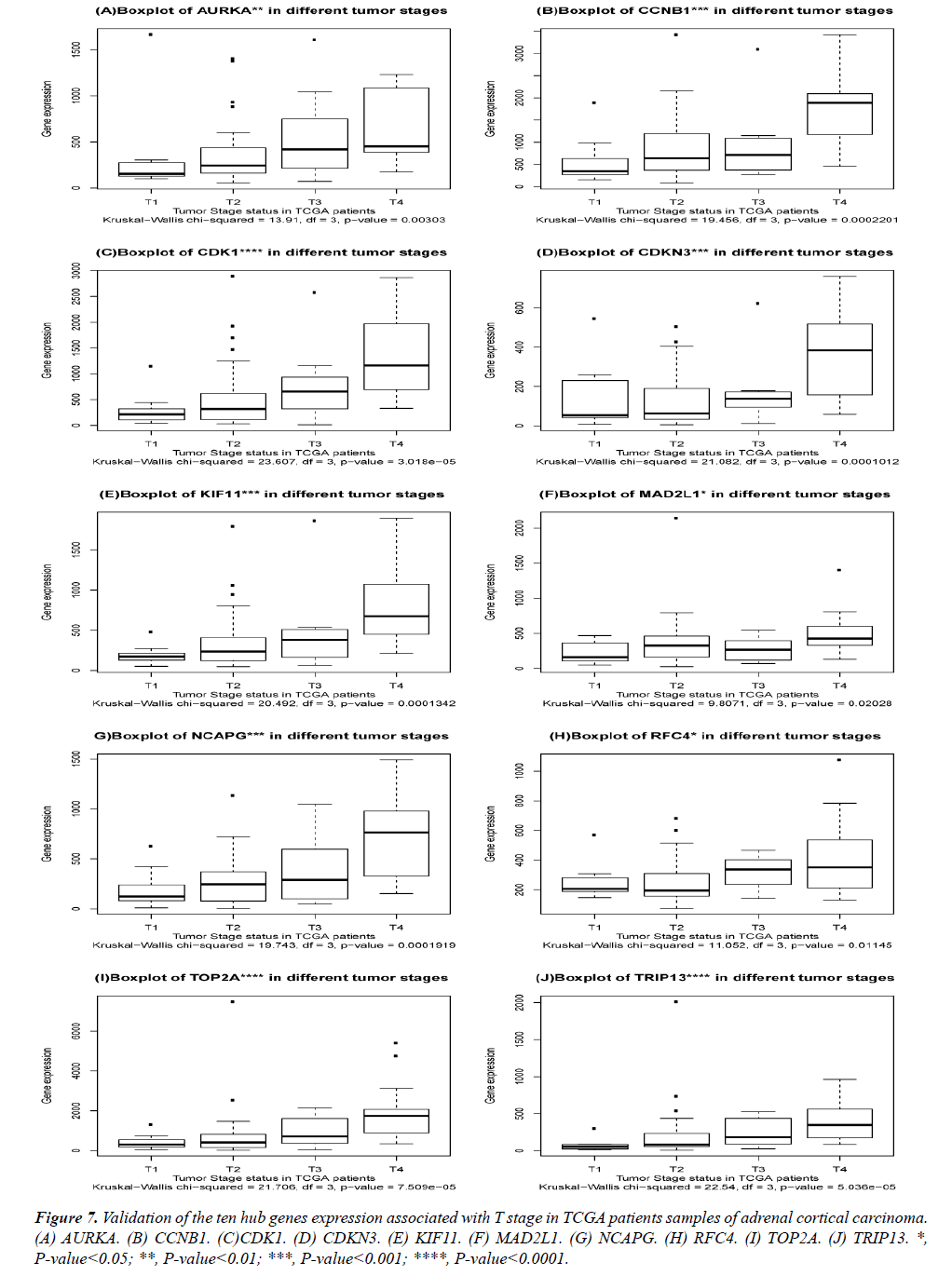
The DEGs of metastatic ACCs were acquired by comparing normal adrenal tissue and primary adrenal cortical carcinoma tissue, and the hub genes were identified by Cytohubba. Those hub genes were then examined by the TCGA datasets. First, these genes expression in M1 and M0 patients were showed in Figure 5. All of them were significant in distinguishing M0 and M1 staging patients. (Wilcoxon test P-value<0.01). They were higher expressed in M1 than that in M0 patients. CDK1, RFC4, and KIF11 were most outstanding among them (P<0.0001). This outcome suggested our analysis was correct and reliable. Then, the N stage was also tested by Wilcoxon test while the T stage was tested by the Kruskal− Wallis test. Their boxplots of these genes were separately shown in Figure 6 and Figure 7. CCNB1, KIF11, MAD2L1, and TRIP13 were statistical meaningful compared in N0 and N1 groups. Others were not related to the N stage. Moreover, the medium expression of most genes grows with the T stage increases. All of them were linked to tumor stages (P<0.05). CDK1, TOP2A and TRIP13 were most significant in different T stage groups (P<0.0001).
Figure 7: Validation of the ten hub genes expression associated with T stage in TCGA patients samples of adrenal cortical carcinoma. (A) AURKA. (B) CCNB1. (C)CDK1. (D) CDKN3. (E) KIF11. (F) MAD2L1. (G) NCAPG. (H) RFC4. (I) TOP2A. (J) TRIP13. *, P-value<0.05; **, P-value<0.01; ***, P-value<0.001; ****, P-value<0.0001.
Discussion
As mentioned above, adrenal cortical carcinoma was a rare disease. It was caused by a cancerous growth in the outer layer of the adrenal glands [23]. Surgery removing the adrenal gland and surrounding tissue was the main method to the disease, and its spreading to other organs and tissues was often lack of therapy methods accompanied with poor prognosis [24]. Discovering the hub biomarkers becomes meaningful for its accuracy therapy [25]. In our research, the microarray GSE90713 including 5 normal and 58 tumor samples were downloaded from the GEO datasets. After being analyzed by BrB-arraytools, a total of 405 DEGs were identified. The upregulated DEGs were mainly involved in the processes of cell division and cell cycle. The down regulated DEGs were mainly involved in the processes of response to hormone and extracellular exosome. The DEGs were inputted into the STRING website, the PPI network was visualized by Cytoscape. The hub genes were identified by Cytohubba, and top 10 genes were obtained: CDK1, RFC4, KIF11, TOP2A, CCNB1, MAD2L1, AURKA, NCAPG, CDKN3, TRIP13. These genes are upregulated in the metastasis ACCs and associated with mitotic cell cycle and so on. In addition, the microRNAs and TFs were also inferred by Webgestalt and their network was shown in Fig 4, including 12TFs and 18 microRNAs. The TFs contained E2F, E2F4DP1, MYCMAX, NFY and so on. MicroRNAs included hsa-mir-492, hsa-mir-488, hsa-mir-34b, hsa-mir-520d, hsamir- 517, hsa-mir-501, hsa-mir-190, hsa-mir-186 and so on. The top ten genes were then validated by the TCGA datasets, which indicated these genes were relative highly expressed in metastasized tumors. Their high expression of these genes indicated poor prognosis of the patients. RFC4 and PCNA were necessary for elongation of DNA templates by DNA polymerase; RFC possessing DNA-dependent ATPase activity were biologically active in various malignant tumors [26,27]. KIF11 was a kinesin-like protein involved in spindle dynamic activity, which had been reported in oral cancer [28,29]. TOP2A was a DNA topoisomerase, regulating the DNA topologic states during transcription. It also amplified in several cancers and its mutation had been connected to the drug resistance [30-32]. CCNB1 was a cell cycle-regulated protein participating in mitosis and formation of MPF [33]. MAD2L1 was involved in the accuracy of chromosome segregation and stop the attack until all chromosomes were correctly positioned at the metaphase plate [34]. CDKN3 was a cyclin-dependent kinase inhibitor and dephosphorylated CDK2 kinase to prevent its activation [35]. CDK1 activated cell transcription factor Hcm1 and targeted it for degradation through phosphorylation of distinct sites, it also played crucial roles in regulating cell cycle progression from G1 to S, through S, and G2 to M phase [36,37]. AURKA was also a cell cycle-regulated kinase participating in microtubule construction and structure steady in mitosis anaphoresis. AURKA was positive in centrosome and spindle poles of different cell mitotic phases [38,39]. NCAPG probably helped introduce positive supercoils into relaxed DNA, Zhang et al. reported that knockdown of NCAPG induces HCC cell mitosis and inhibits cell growth, proliferation, and migration in vitro. TRIP13 interacted specifically with the ligand binding domain and may play a role in early-stage non-small cell lung cancer [40,41]. It was also involved in controlling G2/prophase processes such as DNA break formation and recombination, checkpoint signaling, and chromosome synapsis; it was a conserved member of ATPases associated with diverse cellular activities [42]. These genes of CCNB1, KIF11, MAD2L1, and TRIP13 were closely associated with tumor stage and prognosis, as well as high HR. They may serve as therapeutic targets for the adrenal carcinoma. The Drugbank and upertargets may be used for searching potential drugs for these genes. Their actuals effects on metastasis ACCs need further experimental validations.
Conclusion
In conclusion, CDK1, RFC4, KIF11, TOP2A, CCNB1, MAD2L1, AURKA, NCAPG, CDKN3, TRIP13 have been identified as hub genes in metastasized ACCs. They were not only associated with tumor’s metastasis but also tumor’s staging and patients’ prognosis, except for CCNB1, KIF11, MAD2L1, and TRIP13, others were not meaningful in local lymph node invasion. These genes of CCNB1, KIF11, MAD2L1, and TRIP13 were closely associated with tumor TMN stage and prognosis; they may serve as therapeutic targets for the adrenal carcinoma.
Competing Interests
The authors declare that the research was conducted in the absence of any commercial that could be construed as a potential conflict of interest.
Availability of Data and Materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request. The gene chip GSE90713 is available online.
Consent for Publication
All the authors have consented for the publication.
Authors' Contributions
Di Yu contributed to figure construction and article writing while Chen Dongshan, and YuWei helped with drafting and proofreading, and Yan Lei provided funding and technical support and monitored the whole processes.
Ethics Approval and Consent to Participate
Not applicable.
Funding
This study was supported by financial grants from the Natural Science Foundation of Shandong Province (Grant nos. GG201703180001, and ZR201709230247).
Acknowledgments
Thanks for all the people who help with the analysis.
References
- Else T, Kim AC, Sabolch A, Raymond VM, Kandathil A, Caoili EM, Jolly S, Miller BS, Giordano TJ, Hammer GD. Adrenocortical carcinoma. Endocr Rev 2014; 35: 282-326.
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68: 7-30.
- Rossella Libe. Adrenocortical carcinoma (ACC): diagnosis, prognosis, and treatment. Front Cell Dev Biol 2015; 3: 45.
- Puglisi S, Perotti P, Cosentini D, Roca E, Basile V, Berruti A, Terzolo M. Decision-making for adrenocortical carcinoma: surgical, systemic, and endocrine management options. Expert Rev Anticancer Ther 2018; 18: 125-1133.
- Reimondo G, Puglisi S, Zaggia B, Basile V, Saba L, Perotti P, De Francia S, Volante M, Zatelli MC, Cannavò S, Terzolo M. Effects of mitotane on the hypothalamic-pituitary-adrenal axis in patients with adrenocortical carcinoma. Eur J Endocrinol 2017; 177: 361-367.
- Opden Winkel J, Pfannschmidt J, Muley T, Grünewald C, Dienemann H, Fassnacht M, Allolio B. Metastatic adrenocortical carcinoma: results of 56 pulmonary metastasectomies in 24 patients. Ann Thorac Surg 2011; 92: 1965-70.
- Goel G, Sun W. Cancer immunotherapy in clinical practice -- The past, present, and future. Chin J Cancer 2014; 33: 445-57.
- Barrett T1, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013; 41: 991-995.
- Simon R1, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-array tools. Cancer Inform 2007; 3: 11-17.
- Zhao Y, Simon R. BRB-Array tools data archive for human cancer gene expression: A unique and efficient data sharing resource. Cancer Informatics 2008; 6: 9-15.
- Sun Y1, Li X, Jiang C, Xiao M, Zheng Y, Zhang J, Bu J. Data mining and bioinformatics of the expression data of esophageal squamous cell carcinoma. Cell Biochem Biophys 2014; 69: 481-485.
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4: 44-57.
- Maere S, Heymans K, Kuiper M. BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 2005; 21: 3448-3449.
- Mi H, Huang X, Muruganujan A, Tang H, Mills C, Kang D, Thomas PD. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res 2017; 45: 183-189.
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43: 447-452.
- Shannon P1, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13: 498-504.
- Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. CytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014; 8: 4-11.
- Wang J1, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 2013; 41: 77-83.
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011; 27: 1739-1740.
- Wang JH, Zhao LF, Lin P, Su XR, Chen SJ1, Huang LQ, Wang HF, Zhang H, Hu ZF, Yao KT, Huang ZX. GenCLiP 2.0: A web server for functional clustering of genes and construction of molecular networks based on free terms. Bioinformatics 2014; 30: 2534-2536.
- Cancer Genome Atlas Research Network: Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013; 45: 1113-1120.
- Tang Z1, Li C1, 2, Kang B1, Gao G3, Li C2, 3, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017; 45: 98-102
- Adrenocortical Carcinoma Treatment (PDQ (R)): Health Professional Version BTI - PDQ Cancer Information Summaries 2017.
- Kranjcevic K. [ADRENOCORTICAL CARCINOMA]. Acta Med Croatica 2016; 70: 315-318.
- Mete O, Gucer H, Kefeli M, Asa SL. Diagnostic and prognostic biomarkers of adrenal cortical carcinoma. Am J Surg Pathol 2018 42: 201-213.
- Li Y, Gan S, Ren L, Yuan L, Liu J, Wang W, Wang X, Zhang Y, Jiang J, Zhang F, Qi X. Multifaceted regulation and functions of replication factor C family in human cancers. Am J Cancer Res 2018; 8: 1343-1355.
- Majka J, Burgers PM. The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol 2004; 78: 227-260.
- Mirzaa GM, Enyedi L, Parsons G, Collins S, Medne L, Adams C, Ward T, Davitt B, Bicknese A, Zackai E, Toriello H, Dobyns WB, Christian S. Congenital microcephaly and chorioretinopathy due to de novo heterozygous KIF11 mutations: Five novel mutations and review of the literature. Am J Med Genet A 2014; 164: 2879-2886.
- Daigo K, Takano A, Thang PM, Yoshitake Y, Shinohara M, Tohnai I, Murakami Y, Maegawa J, Daigo Y. Characterization of KIF11 as a novel prognostic biomarker and therapeutic target for oral cancer Int J Oncol, 2018; 52: 55-165.
- Romero A, Martín M, Cheang MC, López García-Asenjo JA, Oliva B, He X, de la Hoya M, García Sáenz JÁ, Arroyo Fernández M, Díaz Rubio E, Perou CM, Caldés Llopis T. Assessment of Topoisomerase II alpha status in breast cancer by quantitative PCR, gene expression microarrays, immunohistochemistry, and fluorescence in situ hybridization. Am J Pathol 2011; 178: 1453-1460.
- Jarvinen TA, Liu. ET. Topoisomerase IIalpha gene (TOP2A) amplification and deletion in cancer--More common than anticipated. Cytopathology 2003; 14: 309-313.
- Bronner C, Hopfner R, Mousli M. Transcriptional regulation of the human topoisomerase IIalpha gene. Anticancer Res 2002; 22: 605-612.
- Wolgemuth DJ. Function of cyclins in regulating the mitotic and meiotic cell cycles in male germ cells. Cell Cycle 2008; 7: 3509-3513.
- Avram S, Mernea M, Mihailescu DF, Seiman CD, Seiman DD, Putz MV. Mitotic checkpoint proteins Mad1 and Mad2 - structural and functional relationship with implication in genetic diseases. Curr Comput Aided Drug Des 2014; 10: 168-181.
- Cress WD, Yu P, Wu J. Expression and alternative splicing of the cyclin-dependent kinase inhibitor-3 gene in human cancer. Int J Biochem Cell Biol 2017; 91: 98-101.
- Benanti JA. Create, activate, destroy, repeat: Cdk1 controls proliferation by limiting transcription factor activity. Curr Genet 2016; 62: 271-276.
- Sakurikar N, Eastman A. Critical reanalysis of the methods that discriminate the activity of CDK2 from CDK1. Cell Cycle 2016; 15: 1184-1188.
- Levinson NM. The multifaceted allosteric regulation of Aurora kinase A. Biochem J 2018; 475: 2025-2042.
- Li M, Gao K, Chu L, Zheng J, Yang J. The role of Aurora-A in cancer stem cells. Int J Biochem Cell Biol 2018; 98: 89-92.
- Zhang Q, Su R, Shan C, Gao C, Wu P. Non-SMC Condensin I Complex, Subunit G (NCAPG) is a Novel Mitotic Gene Required for Hepatocellular Cancer Cell Proliferation and Migration. Oncol Res 2018; 26: 269-276.
- Kang JU, Koo SH, Kwon KC, Park JW, Kim JM. Gain at chromosomal region 5p15.33, containing TERT, is the most frequent genetic event in early stages of non-small cell lung cancer. Cancer Genet Cytogenet 2008; 182: 1-11.
- Vader G. Pch2 (TRIP13): Controlling cell division through regulation of HORMA domains. Chromosoma 2015; 124: 333-339.