ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 20
Microarray analysis of chicken DT40 cells treated with bursopentin
Xiang-Bo Ji1,2#, Jun Luo3#, Xiu-Li Feng1, Qiu-Liang Xu2, Man Teng3, Dong Zhao3, Xin-Feng Li1, Gai-Ping Zhang3 and Pu-Yan Chen1*
1College of Veterinary Medicine, Nanjing Agricultural University, Nanjing, China
2Henan University of Animal Husbandry and Economy, Zhengzhou, China
3Key Laboratory of Animal Immunology, Henan Academy of Agricultural Sciences, Zhengzhou, China
#These authors contributed equally to this work
- *Corresponding Author:
- Pu-Yan Chen
College of Veterinary Medicine
Nanjing Agricultural University, China
Accepted date: July 10, 2017
The Bursa of Fabricius (BF), a central lymphoid organ in birds, is of importance to B cell differentiation and immune modulation. Bursopentin (BP5) is an immune regulatory bioactive peptide derived from BF. In this study, the proliferation and transcriptome profiles of chicken pre-B DT40 cells treated with or without BP5 were investigated by cell proliferation assay and microarray analysis. The results indicated that BP5 showed concentration dependent dual-effects on the proliferation of DT40 cells. According to the results of gene microarray, 3022 genes were differentially expressed. The expression levels of randomly selected differential genes were verified by real time PCR, which were correlated with the microarray results. And the results of GO analysis showed that the differential expression genes were mainly involved in biological processes of innate immune response. Also, KEGG analysis indicates that toll like receptor (TLR) and NF-kappa B signaling pathway may play important roles in DT40 cells treated with BP5. These results will provide a valuable basis for understanding the potential molecular mechanism of BP5 on B lymphocyte.
Keywords
Bursopentin, Bursa of fabricius, Gene microarray, Pre-B lymphocyte DT40 cell, Transcriptome
Introduction
The avian primary lymphoid organ Bursa of Fabricius (BF), sometimes also called “the cloacal thymus”, plays a very important role in the proliferation and differentiation of antibody producing B lymphocytes [1]. Previous studies have demonstrated that certain peptides that have been extracted from the BF, e.g., bursin and bursal anti-steroidogenic peptide, are able to stimulate specific immune cell subsets [2,3]. BF extracts containing multiple biologically active factors have been used for the majority of such studies [4-6] that did, however, reveal interesting effects on avian and mammal humoral immune systems. They have been shown to modulate B cell proliferation and antibody production in mice or chicks.
Recently, Li et al. isolated a new pentapeptide from BF extracts of chicks, named bursopentin (Cys-Lys-Arg-Val-Tyr) [7]. Several studies have reported BP5 to exert a variety of functions. For example, it has been found to enhance antibody production, to stimulate the proliferation of splenic T cells and B lymphocyte in mice, and to augment the cytotoxic activity of activated splenocytes cytotoxic T lymphocyte against NIH3T3 cells [7]. Furthermore, BP5 seems to protect murine peritoneal macrophages from oxidative stress induced by LPS [8], and to increase the number of CFU-pre B [9]. Also BP5 regulated the redox homeostasis of WEHI-231 cells, modulated the immune function of DCs [10] and protected DCs from oxidative stress [11]. However, the mechanism of actions of the immunomodulatory peptide is largely unknown.
The chicken pre-B cell line DT40 expresses immunoglobulin M (IgM) isotype B cell reporter in the plasma membrane [12,13]. Several previous studies provide evidence that supports the hypothesis that DT40 cells may be considered a bursal stem cell line [14,15]. For this reason, we decided to use it for an in vitro investigation of the immunomodulatory functions of BP5.
As far as we know, there are currently few reports concerning the effect and molecular mechanism of BP5 on DT40 cells. We therefore aimed at evaluating the effect of BP5 on the proliferation of DT40 cells. To further understand the molecular mechanism of action of BP5, the transcriptome profiles of DT40 cells treated with BP5 were obtained by microarray analyses and compared with negative controls. Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways and Gene Ontology (GO) analyses were also performed to elucidate the potential molecular mechanism behind BP5-mediated effects on pre-B cells.
Materials and Methods
BP5 was synthesized by Shanghai Sangon Bioscience Co. Ltd (Shanghai, China). The purity of the synthetic BP5 was >99%, as has been proved by reverse phase high performance liquid chromatography. The chicken DT40 cell line (ATCC: CRL2111) was cultured in RPMI 1640 medium (Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) and 5% chicken serum (CHS, Gibco), 50 μM β-Mercaptoethanol, 100 IU/ml penicillin and streptomycin. The cells were incubated in an atmosphere containing 5% carbon dioxide at 37°C with relative humidity.
DT40 cells were allowed to grow until reaching a concentration of 0.4-0.6 × 106 cells/ml. Cells were then collected and washed with RPMI medium for three times. After being transferred to RPMI medium with 1% FBS and 1% CHS, cells were seeded into tissue culture flasks at densities of 1 × 106 cells/ml and were treated with 0.02, 0.2, 2, and 20 μg/ml BP5, respectively. Cells were collected from 24 h to 96 h post seeding every 24 h and quantified using a hemocytometer under a light microscope (Olympus). Cell viability was assessed by Trypan blue exclusion test. All experiments above were carried out in triplicate.
DT40 cell line (1 × 106 cells/ml) were treated with 0.02 μg/mL BP5 (n=3) or PBS (n=3) for 4 h, subsequently, cells were collected for total RNA isolation using the TRIzol reagent (Invitrogen, USA) following the manufacturers’ instructions. Cells treated with PBS were used as control. The total RNA of each sample was tested by the NanoDrop One (Thermo, USA) and the RNA integrity was checked by the agarose gel electrophoresis. And the total RNAs with OD260/OD280 ≥ 1.8 and RNA integrity number ≥ 8.0 were used for the microarray analysis.
The microarray analysis of RNA extracted from BP5 (0.02 μg/ml) treatment and PBS control group was used by the Agilent Array platform. In brief, total RNA (1 μg) of each sample was processed to fluorescent cRNA following the manufacturer’s standard protocol (Version 5.7, Agilent Technologies). All the labeled samples were hybridized by the standard Agilent Microarray Expression Analysis protocol (Agilent, USA). The results of microarray data were submitted to a Gene Expression Omnibus database (accession number: GSE30396).
The results were output using Agilent Feature Extraction software and analysed by the Agilent GeneSpring GX software. Gene expression was considered to be affected by BP5 treatment, if fold changes of ≥ 1.5 were measured.
In order to analyse the expression of gene-specific mRNA, the DT40 cell line was incubated in standard conditions and the total RNA was extracted as stated above. Total RNA was then subjected to RT-PCR using a SYBR Green PrimeScript RTPCR Kit (Takara, Dalian). Gene expressions were determined using a set of 12 target specific primers (Table 1) for genes: CSNK2A1, FGF3, FGF8, FZR1, IRF7, MAP3K7, MYD88, RAC1, RAP1B, SKP1, TCEB1 and TRIP12. And the levels of β-actin transcripts were used as an internal reference (Table 1).
| Gene | Primer name | Primer sequence |
|---|---|---|
| β-actin | Reverse | TTACTCCCACAGCCAGCCAT |
| Forward | TAGCTGTCTTTCTGGCCCAT | |
| RAP1B | Reverse | CAAGCTAGTGGTTCTTGGTTCTGGA |
| Forward | GCATACACTGTTGCGCATCTACTTC | |
| FZR1 | Reverse | TCCTGGGAGCAGGGATTGAG |
| Forward | GACTGGAACGTTTGGTGCTGAG | |
| TCEB1 | Forward | AGAGCATGCATTAACATCAGGAACA |
| Reverse | GGTGCAATTGGGAATTCAGGA | |
| CSNK2A1 | Forward | CTCCTGTCAGCAGTGCGAGTATG |
| Reverse | ATGACGGGTGAGCCTGCTAGA | |
| FGF3 | Reverse | TGTCGGGATCGTCGCTATCA |
| Forward | GGTACAGACGGGATGCATAGGTG | |
| MAP3K7 | Forward | CGCCTCCGCTGAAATGATTG |
| Reverse | ATGAGGCAGAGGTTCAGCAC | |
| MYD88 | Forward | AATGGACACTGAGCTCTGCC |
| Reverse | CAAACCCGATCTGTGGGACA | |
| RAC1 | Forward | TTCCATGGTTTCGTTTGTTTGA |
| Reverse | TTGCAACCAAGCCCTTACCA | |
| TRIP12 | Forward | ACCGGCCTAATAACAATCCAGG |
| Reverse | AACTACAGCCTCTTCCTCCTGCT | |
| FGF8 | Reverse | TCATCGTCGAGACCGACACC |
| Forward | CAGTCCTTGCCTTTGCCGTTA | |
| SKP1 | Reverse | TTAAGCTGCAGAGTTCAGATGGAGA |
| Forward | CTGGGTCATCATCGCCTTCA | |
| IRF7 | Reverse | TGAAGGTCAACACACCACAGGAG |
| Forward | CCCAACCACAAAGCTTATTGCAG |
Table 1. The primer sequence of RT-PCR.
The Gene Ontology (GO) of differentially expressed genes were functionally analysed according to the GO website (www.geneontology.org), that includes three components of defined terms to tell the attributes of gene product. KEGG signaling pathway analysis was performed to all the differentially expressed genes basing on the latest NCBI, KEGG, Biocarta and Reactome database.
All the data are presented as means ± standard deviations from at least three independent experiments. Data were tested with analyses of variance (one-way or two-way ANOVA). P values<0.05 were considered to be statistically significant.
Results
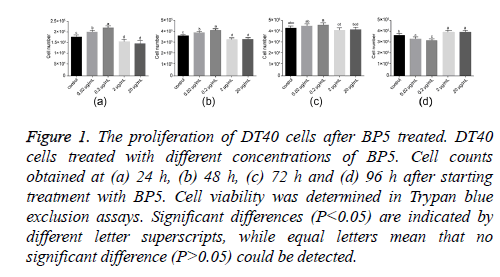
To study the effects of BP5 on DT40 cell proliferation, the numbers of DT40 cells treated with different concentrations of BP5 (0.02-20 μg/ml) were determined at 24, 48, 72 and 96 h post treatment. As shown in Figure 1, BP5 treatment resulted in dual effects on DT40 cell proliferation. BP5 stimulated the proliferation of DT40 cell by concentration dependent. Cell numbers were significantly augmented after cells had been exposed to 0.02 and 0.2 μg/ml BP5 for 24 and 48 h. This trend, however, was interrupted when BP5 was applied in concentrations exceeding 2 μg/ml. Here, cell counts dropped below control values. After prolonged exposure to BP5, cell viability started to diminish. Then the live cell populations of 0.02 and 0.2 μg/ml BP5 treatments decreased rapidly, meanwhile conversely the cell numbers of 2 and 20 μg/ml BP5-treated decreased slowly at 72 and 96 h. These data indicate that BP5 displays dual effects on the proliferation of DT40 cell by concentration dependent.
Figure 1: The proliferation of DT40 cells after BP5 treated. DT40 cells treated with different concentrations of BP5. Cell counts obtained at (a) 24 h, (b) 48 h, (c) 72 h and (d) 96 h after starting treatment with BP5. Cell viability was determined in Trypan blue exclusion assays. Significant differences (P<0.05) are indicated by different letter superscripts, while equal letters mean that no significant difference (P>0.05) could be detected.
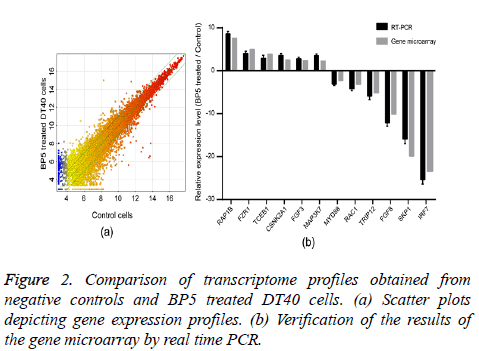
In order to identify the molecular mechanism of BP5 on pre-B lymphocytes, the transcriptome profiles of DT40 cells incubated for 4 h with 0.02 μg/ml BP5 and negative controls were determined by gene microarray analysis. Scatter plots enabled us to identify genes whose expression underwent at least a 1.5 fold change upon treatment with BP5 (Figure 2a). The analysis of transcriptome profiles showed that there were 3163 genes up regulated, while there were 3539 genes down regulated after BP5 treatment (Supplement Table S1). The expression levels of 12 selected genes were verified by a real time PCR approach and correlated with gene microarray results (Figure 2b).
GO analysis was carried out to identify the obviously changed biological functions of those genes found to be differentially expressed in DT40 following BP5 treated. The results showed that more than 1000 differentially expressed genes of all significantly regulated genes were involved in GO terms for Biological Processes (BP), Molecular Function (MF) and Cellular Component (CC) (supplement Table S2). GO term of biological processes (P value<0.05) affected by the differentially expressed genes were mostly associated with immune response, B cell biological processes, immunoglobulin secretion and cytokine production.
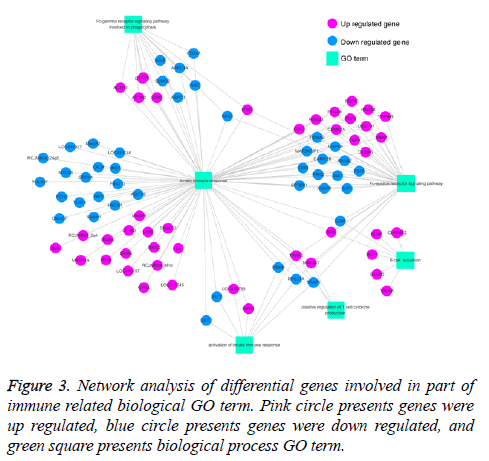
Previous study showed that BP5 exhibited pleiotropic actives in immune response in mice, such as enhancing antibody production, promoted lymphocyte proliferation [7]. In our current study, the results showed that genes that significantly regulated were involved in lots of immune related biological processes in DT40 cells in response to BP5 (Supplement Table S3). To clarify the interaction of differential expression genes related with biological processes, genes related with 6 immune-related biological processes terms were selected, and GO-Gene-Network was made (Figure 3). As shown in Figure 3, 81 differentially expressed genes were involved in innate immune response (GO: 0045087), in which the up regulated genes TRAF2, MAP3K7 were also involved in positive regulation of T cell cytokine production (GO: 0032481) and Fc-epsilon receptor signaling pathway (GO: 0038095). Also, the down regulated genes CD86, IKBKB, RAC1 and CD80 were involved in B cell activation (GO: 0042113), activation of innate immune response (GO: 0002218), positive regulation of T cell cytokine production and Fc epsilon receptor signaling pathway.
Cytokines are important mediators in cell induced immune response [16]. Previous studies showed that BP5 could remarkably induce both Th1 and Th2 cytokines secretion, which indicated that BP5 might have the function of regulating T cell mediated immune responses by balancing Th1 and Th2 responses [7]. In this paper, except for mechanism of the effect of BP5 on immune response in DT40 cell, we also determined intracellular transcriptional changes that related to cytokine production and regulation. As shown in Table 2, there were 15 up regulated genes and 17 down regulated genes, which were involved in 8 cytokine production and regulation biological processes, such as positive regulation of interleukin-8 secretion (GO: 2000484), positive regulation of type I interferon production (GO: 0032481) and positive regulation of cytokinesis (GO: 0032467).
| GO ID | GO Term | P value | Up regulated genes | Down regulated genes |
|---|---|---|---|---|
| GO:2000484 | Positive regulation of interleukin-8 secretion | 0.00465 | LOC416197//HYAL2 | FCN2//WNT5A |
| GO:0032481 | Positive regulation of type I interferon production | 0.00762 | RIPK1//MRE11A | IKBKB//RPS27A//CRCP// |
| O\POLR2F//IRF7//MYD88//LOC418927//IRF1 | ||||
| GO:0032743 | Positive regulation of interleukin-2 production | 0.00804 | MAP3K7//TRAF2 | TRAF6//CD83//PDE4B |
| GO:2000778 | Positive regulation of interleukin-6 secretion | 0.0235 | LOC416197// | |
| HYAL2//XBP1 | ||||
| GO:0045086 | Positive regulation of interleukin-2 biosynthetic process | 0.03001 | GLMN//IRF4 | CD86//CD80 |
| GO:0045351 | Type I interferon biosynthetic process | 0.03693 | IRF7//MYD88 | |
| GO:0045404 | positive regulation of interleukin-4 biosynthetic process | 0.03693 | IRF4 | CD86 |
| GO:0032467 | Positive regulation of cytokinesis | 0.01772 | SVIL//SPAST//PKN2//CXCR5//OPN1LW//CUL3 | KIF4//CIT |
Table 2. Profile of cytokine related biological processes by BP5 treatment (Fold ≥1.5).
The present results show that 38 GO term of cellular component and 59 GO terms of molecular functions were significantly regulated (Supplement Table S2). In the molecular function component, genes such as TRAF6, TRAF2, CD80, CD86, RAC1 and IRF7 were involved in protein binding (GO: 0005515) and ubiquitin protein ligase binding (GO: 0031625). In the cellular component ontology, TRAF6, TRAF2 and IKBKB were involved in CD40 receptor complex (GO: 0035631).
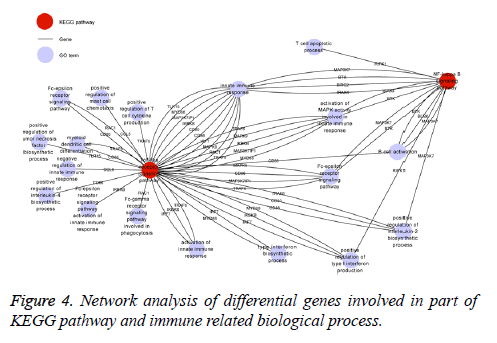
The KEGG database was used to identify those signaling pathways influenced by differentially expressed genes (fold change>1.5). According to the results, there were 26 signaling pathways modulated by BP5 (p<0.05) which were involved in cell signal transduction, pathogens infection, and cancer related signaling pathways (Supplement Table S4). Consistent with results from the biological processes of GO analysis of differential expression genes, most of genes involved in tolllike receptor (TLR) and NF-kappa B signaling pathway are associated with pre-B lymphocyte development and humoral immunity as can be seen in Figure 4.
Discussion
The BF is an organ of utmost importance for the humoral immune system of birds. B cell differentiation and antibody production highly depend on its function. It has been reported that chicks do not respond to antigen stimulation when their BF is removed. Antibody function could, however, be recovered after administration of BF extracts [17-19]. Therefore, BF peptides have gained a lot of attention in recent years. There are a lot of studies suggesting that these peptides take very important part in basic cellular processes of B cells [12,20]. A previous study has proven that BP5 derived from the BF is an immune competence factor, which up regulates the process of antibody production in mice [7]. Recent reports further proposed that BP5 may regulate various cellular processes of WEHI-231 B cells [9], and that it may have immune pharmacological potential by suppressing the secretion of anti-inflammatory and pro-inflammatory cytokines of DCs in mice [11,21].
In the present study, we evaluated the effects of BP5 exerted on the proliferation of chick pre-B DT40 cells. We found that BP5 mediated concentration dependent dual effects in this cell line. Lower concentrations of BP5 induced cell proliferation, while higher concentrations of BP5 inhibited it. It was reported that another peptide BPP II derived from BF exerted the similar effects on DT40 cells [20].
In order to further illuminate the molecular mechanism of BP5 in avian B cells, the transcriptome profile of DT40 cells treated with 0.02 μg/ml BP5 was compared to the untreated cells.
GO analysis showed most differential genes were related with innate immune responses, B cell activation, immunoglobulin secretion and regulation of cytokines production, which were significantly over or down represented in BP5 treated DT40 cell. And GO-gene-network identified many regulated genes, such as TRAF6, TRAF2, CD80, and CD86 were taken part in diverse immune related GO terms of biological processes, which indicated the mutual regulated effects among hummoral response, cellular response immunity, and cytokines production. In addition, genes taking part in B cell related biological processes were selected to explore the molecular mechanism of BP5 on B cell development and antibody production.
It was reported that Bcl-2 played a vital role in modulating the cell cycle of many cells including B cell, which lead to effect activation of B cell by apoptosis [22]. Our results indicated that the expression of Bcl-2-13 was down regulated in DT40 cells treated with BP5. During the initial stage of B cell activation process, a number of different kinases are essential in signal transduction pathways to magnitude the signal by BCR [23,24]. It was reported that Blk belonging to receptor associated PTKs could be activated by ITAM and then induces down-stream signal transduction pathways. It was also found that the biological activation of BP5 could be reduced by PTK inhibitor [7]. In our study, except for gene of Blk, some key signal transducer genes, such as the Btk and BLNK which were involved in the signal pathway of B cell activation [25] were up regulated. And it has been reported that Btk plays important roles in BCR signaling and function in cell proliferation and cell survival in B cell malignancies [26], while BLNK bridges the B cell receptor associated kinases with a lot of pathways and may modulate downstream biological events of B cell [27]. The results indicated that Bcl-2, Blk, Btk, and BLNK might be the signal transcription factors following BP5 treatment in DT40 cells.
Many studies have indicated that B lymphocytes not only display important roles in adaptive immune response, but also contribute to antigen presentation and cytokine production [28]. Our study showed the altered gene expression profiles induced by BP5 treatment in DT40 cells are highly related to the innate immune response. Accordingly, we selected some key genes involved in numerous biological processes to explore the mechanism of BP5 on immune responses. It has been reported that TRAFs play important roles in innate receptor signaling [29]. TRAF2 and TRAF6 are members of TRAF super family, which takes important part in cell proliferation, differentiation and apoptosis that associated with innate immunity and anti-inflammatory [30]. CD40 is a number of TNF family of cell surface proteins and costimulatory protein for the activation of Antigen Presenting Cell (APC) [31]. The activation of APC and induction of downstream effects requires the binding of CD154 on TH cell to CD40 [32]. According to cellular component ontology, TRAF6 and TRAF2 were involved in CD40 receptor complex, which is an essential factor for B cells activation. Expression of TRAF2 involved in activation of innate immune response and myeloid dendritic cell differentiation was elevated in BP5 treated DT40 cell, whereas TRAF6 which present important mediators of innate receptor signaling was down regulated. These findings indicated that BP5 might regulate either innate immunity or adaptive immunity through altering the expression level of the converged signal molecules.
As previously reported that BP5 could remarkably stimulate both IFN-γ and IL-4 production in vivo [7]. To study how BP5 signaling converges on cytokine secretion, we briefly consider the roles of various genes and their possible roles in regulation of cytokine production. Except for TRAF2 and TRAF6, genes of some regulatory molecules which were involved in B cell mediated immunity and cytokine secretion were regulated. It was reported that CD80 and CD86 were presented on APC and they can induce TH cells by interacting with CD28 and CTLA-4 [33]. We found that both of these genes were down regulated and both involved in biological processes of regulation of IL2 and IL4 biosynthesis. The IRF7 gene, which has recently been identified as the key regulator of type I IFN activation [34] and TNFSF13B, a number of TNF ligand and receptor super family, which acts as a potent B cell activator and also involved in cytokine secretion biological processes [35], were down regulated. These results explained the molecular mechanism of effects of BP5 on cytokines production.
It is well known that B cell development inexorably rely on the signal transductions and networks of various signaling pathway [36,37]. Considering the fact that 0.02 μg/ml BP5 induced proliferation of DT40 cells, we predict that there may be multiple transcriptional changes caused by BP5 involved in various signal pathways. The results of KEGG pathway analyses showed that there were 26 signaling pathways affected by BP5 treatment. TLR and NF-κB signaling pathways associated with immunity and cell development are focused on due to the fact that BP5 has been shown to exert dominant effects on immune responses [7,9,11]. It has been reported that TLR signaling pathway plays an important role in innate immune response, which may occur by MyD88 dependent and MyD88 independent manner [38,39]. TLRmediated intracellular signal transduction utilizes the signaling molecule MyD88 and does also involve the nuclear translocation of NFκB, the activation of protein kinase B/Akt, and MAPK, and the phosphorylation of the IRF transcription factor family [40]. In the present study, MyD88 which involved in biological processes of positive regulation of type I interferon production was found to be down regulated.
IRAKs are important mediators of TLR induced intracellular signal transduction as they may potentiate the down steam events [41]. Some studies indicated that IRAK4 and IRAK1 were phosphorylated and dissociated from MyD88 upon stimulation that results in the activation of TRAF6 [42-45]. Our results showed that IRAK4 and IFN-β were up regulated, while TRAF6 was down regulated in DT40 cells treated with BP5. Except for that the IRF7 gene involved in TLR signaling pathway, which has recently been identified as the key regulator of type I IFN activation [34], was also down regulated. These results suggested that BP5 maybe balance the innate immune responses through down regulating MyD88 to induce downstream intracellular signal transduction.
It was also reported that NF-kappa B signaling pathways mediated multiple aspects in innate and adaptive immune responses through TLR signaling pathways [46,47]. The KEGG pathway data showed that NF-kappa B signaling pathway was significantly regulated, in which genes of Btk, BLNK, IRAK4, CSNK2A1, and MAP3K7 were up regulated. And according to GO analysis, all the genes mentioned above were associated with biological processes of innate immune response, B cell activation, Fc-epsilon receptor signaling pathway and positive regulation of T cell cytokine production. These results indicated that NF-kappa B signaling pathways may take an important role in DT40 cells treated with BP5.
In summary, our studies indicate that BP5 exerts a strong regulating effect on DT40 cell biological processes. The results of our gene microarray analysis, the transcriptome profiles of cells treated with BP5 and untreated controls, enable us to speculate on the signaling pathways that mediate the effects of BP5 on B cell development. In our subsequent study, the key identified regulatory genes would be focused further to study the molecular mechanism of BP5 induced effects.
References
- Audhya T, Kroon D, Heavner G, Viamontes G, Goldstein G. Tripeptide structure of bursin, a selective B-cell-differentiating hormone of the bursa of fabricius. Science 1986; 231: 997-999.
- Byrd JA, Hayes TK, Wright MS, Dean CE, Hargis BM. Detection and partial characterization of an anti-steroidogenic peptide from the humoral immune system of the chicken. Life Sci 1993; 52: 1195-1207.
- Feng XL, Liu QT, Cao RB, Zhou B, Wang FQ, Deng WL, Qiu YF, Zhang Y, Ishag H, Ma ZY, Zheng QS, Chen PY. A bursal pentapeptide (BPP-I), a novel bursal-derived peptide, exhibits antiproliferation of tumor cell and immunomodulator activity. Amino Acids 2012; 42: 2215-2222.
- Baba TW, Humphries EH. Selective integration of avian leukosis virus in different hematopoietic tissues. Virology 1986; 155: 557-566.
- Feng XL, Liu QT, Cao RB, Zhou B, Ma ZY, Deng WL, Wei JC, Qiu YF, Wang FQ, Gu JY, Wang FJ, Zheng QS, Ishag H, Chen PY. Identification and characterization of novel immunomodulatory bursal-derived pentapeptide-II (BPP-II). J Biol Chem 2012; 287: 3798-3807.
- Feng XL, Liu QT, Cao RB, Zhou B, Zhang YP, Liu K, Liu XD, Wei JC, Li XF, Chen PY. Characterization and immunomodulatory function comparison of various bursal-derived peptides isolated from the humoral central immune organ. Peptides 2012; 33: 258-264.
- Li DY, Geng ZR, Zhu HF, Wang C, Miao DN, Chen PY. Immunomodulatory activities of a new pentapeptide (Bursopentin) from the chicken bursa of Fabricius. Amino Acids 2011; 40: 505-515.
- Li DY, Xue MY, Geng ZR, Chen PY. The suppressive effects of Bursopentine (BP5) on oxidative stress and NF-kB activation in lipopolysaccharide-activated murine peritoneal macrophages. Cell Physiol Biochem 2012; 29: 9-20.
- Liu XD, Zhou B, Cao RB, Feng XL, Ma ZY, Chen PY. BP5 regulated B cell development promoting anti-oxidant defence. Amino Acids 2014; 46: 209-222.
- Peng X, Yang L, Chang H, Dai G, Wang F, Duan X, Guo L, Zhang Y, Chen G. Wnt/β-catenin signaling regulates the proliferation and differentiation of mesenchymal progenitor cells through the p53 pathway. PLOS One 2014; 9: 97283.
- Qin T, Yin Y, Yu Q, Yang Q. Bursopentin (BP5) protects dendritic cells from lipopolysaccharide-induced oxidative stress for immunosuppression. PLoS One 2015; 10: 0117477.
- Buerstedde JM, Reynaud CA, Humphries EH, Olson W, Ewert DL. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J 1990; 9: 921-927.
- Gerondakis S, Grumont RJ, Banerjee A. Regulating B-cell activation and survival in response to TLR signals. Immunol Cell Biol 2007; 85: 471-475.
- Bonet C, Giuliano S, Ohanna M, Bille K, Allegra M, Lacour JP, Bahadoran P, Rocchi S, Ballotti R, Bertolotto C. Aurora B is regulated by the mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathway and is a valuable potential target in melanoma cells. J Biol Chem 2012; 287: 29887-29898.
- Lopes-Carvalho T, Kearney JF. Development and selection of marginal zone B cells. Immunol Rev 2004; 197: 192-205.
- Yoshimura A, Naka T, Kubo M. SOCS proteins, cytokine signalling and immune regulation. Nat Rev Immunol 2007; 7: 454-465.
- Baba T, Kita M. Effect of extracts of the bursa of Fabricius on IgG antibody production in hormonally bursectomized chickens. Immunology 1977; 32: 271-274.
- Heller DE, Friedman AR. The effect of crude bursa of Fabricius extracts on the humoral immune response and its recovery in bursectomized chickens. Dev Comp Immunol 1979; 3: 667-681.
- Pisarevskaia LI, Kuznik BI, Tsybikov NN. Role of the bursa of Fabricius in regulating the hemostatic system in the chicken. Fiziol Zh SSSR Im I M Sechenova 1984; 70: 23-31.
- Feng X, Cao R, Zhou B, Liu Q, Liu K. The potential mechanism of Bursal-derived BPP-II on the antibody production and avian pre-B cell. Vaccine 2013; 31: 1535-1539.
- Yin Y, Qin T, Yu Q, Yang Q. Bursopentin (BP5) from chicken bursa of fabricius attenuates the immune function of dendritic cells. Amino Acids 2014; 46: 1763-1774.
- Muscarella DE, Bloom SE. The contribution of c-Jun N-terminal kinase activation and subsequent Bcl-2 phosphorylation to apoptosis induction in human B-cells is dependent on the mode of action of specific stresses. Toxicol Appl Pharmacol 2008; 228: 93-104.
- Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science 2002; 298: 2199-2202.
- Crotty S, Ahmed R. Immunological memory in humans. Semin Immunol 2004; 16: 197-203.
- Tsukada S, Baba Y, Watanabe D. Btk and BLNK in B cell development. Adv Immunol 2001; 77: 123-162.
- Hendriks RW, Yuvaraj S, Kil LP. Targeting Brutons tyrosine kinase in B cell malignancies. Nat Rev Cancer 2014; 14: 219-232.
- Young RM, Staudt LM. Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat Rev Drug Discov 2013; 12: 229-243.
- Stanic B, van de Veen W, Wirz OF, Ruckert B, Morita H, Sollner S, Akdis CA, Akdis M. IL-10-overexpressing B cells regulate innate and adaptive immune responses. J Allergy Clin Immunol 2015; 135: 771-780, e778.
- Chung JY, Park YC, Ye H, Wu H. All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J Cell Sci 2002; 115: 679-688.
- Jin J, Xiao Y, Hu H, Zou Q, Li Y, Gao Y, Ge W, Cheng X, Sun SC. Proinflammatory TLR signalling is regulated by a TRAF2-dependent proteolysis mechanism in macrophages. Nat Commun 2015; 6: 5930.
- Pype S, Declercq W, Ibrahimi A, Michiels C, Van Rietschoten JGI, Dewulf N, de Boer M, Vandenabeele P, Huylebroeck D, Remacle JE. TTRAP, a novel protein that associates with CD40, Tumor Necrosis Factor (TNF) receptor-75 and TNF Receptor-associated Factors (TRAFs), and that inhibits nuclear factor-κB activation. J Biol Chem 2000; 275: 18586-18593.
- Gaspal FM, McConnell FM, Kim MY, Gray D, Kosco-Vilbois MH, Raykundalia CR, Botto M, Lane PJ. The generation of thymus-independent germinal centers depends on CD40 but not on CD154, the T cell-derived CD40-ligand. Eur J Immunol 2006; 36: 1665-1673.
- Ellis JH, Burden MN, Vinogradov DV, Linge C, Crowe JS. Interactions of CD80 and CD86 with CD28 and CTLA4. J Immunol 1996; 156: 2700-2709.
- Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun 2011; 12: 399-414.
- Almaden JV, Liu YC, Yang E, Otero DC, Birnbaum H, Davis-Turak J, Asagiri M, David M, Goldrath AW, Hoffmann A. B-cell survival and development controlled by the coordination of NF-kappaB family members RelB and cRel. Blood 2016; 127: 1276-1286.
- Pieper K, Grimbacher B, Eibel H. B-cell biology and development. J Allergy Clin Immunol 2013; 131: 959-971.
- Reth M, Nielsen P. Signaling circuits in early B-cell development. Adv Immunol 2014; 122: 129-175.
- Banerjee A, Gerondakis S. Coordinating TLR-activated signaling pathways in cells of the immune system. Immunol Cell Biol 2007; 85: 420-424.
- Hua Z, Hou B. TLR signaling in B-cell development and activation. Cell Mol Immunol 2013; 10: 103-106.
- Kawai T, Akira S. TLR signaling. Cell Death Differ 2006; 13: 816-825.
- Conze DB, Wu CJ, Thomas JA, Landstrom A, Ashwell JD. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-κB activation. Mol Cell Biol 2008; 28: 3538-3547.
- Arsura M, Hofmann CS, Golay J, Introna M, Sonenshein GE. A-myb rescues murine B-cell lymphomas from IgM-receptor-mediated apoptosis through c-myc transcriptional regulation. Blood 2000; 96: 1013-1020.
- Kawagoe T, Sato S, Jung A, Yamamoto M, Matsui K, Kato H, Uematsu S, Takeuchi O, Akira S. Essential role of IRAK-4 protein and its kinase activity in Toll-like receptor-mediated immune responses but not in TCR signaling. J Exp Med 2007; 204: 1013-1024.
- Mayumi M, Ohshima Y, Hata D, Kim KM, Heike T, Katamura K, Furusho K. IgM-mediated B cell apoptosis. Crit Rev Immunol 1995; 15: 3-4.
- Wang Z, Wesche H, Stevens T, Walker N, Yeh WC. IRAK-4 inhibitors for inflammation. Curr Top Med Chem 2009; 9: 724-737.
- Bonizzi G, Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol 2004; 25: 280-288.
- Silverman N, Maniatis T. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev 2001; 15: 2321-2342.