ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2015) Volume 26, Issue 4
Impact of G406S and G420R mutants associated with Blood Coagulation Factor Xa: Molecular simulation approach
Shabana kouser Ali, George Priya Doss. C*, Anbalagan M
Medical Biotechnology Division, School of Biosciences and Technology, VIT University, Vellore, Tamil Nadu, India.
- Corresponding Author:
- George Priya Doss. C
Medical Biotechnology Division School of Biosciences and Technology VIT University, Vellore Tamil Nadu, India.
Accepted date: August 04 2015
Protein flexibility and conformational changes are known to be key characteristic for the function of many proteins. Factor Xa is a serine protease which activates thrombin and plays a significant role in blood coagulation pathway. Mutations in Factor Xa could be interfaced with thrombosis. To address this issue, we implemented in silico prediction algorithms SIFT, PolyPhen 2.0, I-Mutant 3.0, Align GVGD, to classify the variants as pathogenic. Based on prediction algorithm scores, two variants G406S and G420R were predicted as highly deleterious from a pool of 58 nsSNPs/variants. To analyze the dynamic behavior mutant proteins G406S and G420R, molecular simulation analysis was carried out for 5000ps. This computational work might help us in understanding the effects of mutations on wild Factor Xa protein
Keywords
Factor Xa, Coagulation, Mutation, SNPs, Molecular simulation
Introduction
Factor X is a serine endopeptidase enzyme, also known as prothrombinase, thrombokinase of the coagulation cascade. Factor X is activated into Xa by Factor IX and Factor VII with its cofactor and tissue Factors. Factor X is the starting Factor of the final pathway or also known as thrombin pathway [1]. It acts by cleaving prothrombin at two different sites (arg-thr and arg-ile bond) which results in the active thrombin. Factor Xa is the activated form of coagulation Factor thrombokinase, also known as a stuartprower Factor. The chromosomal position for human Factor Xa gene is 13q34. Varying in severity of Factor Xa deficiency rare bleeding disorder causes nosebleeds, bleeding of gums, hematuria, menorrhagia, hemarthrosis, intracranial, and pulmonary hemorrhage. Mutations in Factor Xa gene result in deficiency known as congenital Factor Xa deficiency. Single nucleotide polymorphism (SNPs) is the class of variation in human genome. Among them, non-synonymous SNPs (nsSNPs) bring about the progression of amino acid deposits. Therefore, predicting the impact of nsSNPs/variants in coding region by means of computational analysis provides an exceptional way to evaluate the relationship between functional nsSNPs and their disease susceptibility. In light of in silico algothrims we used methods such as SIFT (Sorting Intolerant from Tolerant) [], PolyPhen2.0 [], I-Mutant 3.0 [], and Align GVGD to analyse and predict the impact of mutation in Factor Xa gene. The literature survey on Factor Xa is considerable – a search for article for ‘Factor Xa’ results for many hits, with main contributions from experimental studies [5-7] and few from theoretical studies [8-10]. Despite the vast information, there are no studies of uncomplexed Factor Xa, focusing on molecular dynamics simulation analysis. Henceforth, to develop a coherent approach, we applied computational insight on predicted mutations G406S (184) and G420R (198) with the aid of molecular dynamics simulation using GROMACS [] for 5000ps each. A structural insight to analyze the protein stability in the both wild and mutant structures were analyzed using RMSD (root mean square deviation) and RMSF (root mean square fluctuation).
Materials and Methods
For analysis, nsSNPs related dataset for Factor Xa gene was obtained from National Center for Biotechnology Information (NCBI) and UniPort database. The 3D structure for Factor Xa with PDBID:2VH6 was obtained from Protein data bank [11]. For identifying the deleterious nsSNPs, we used sequence based approach SIFT [12] and sequence and structural based approach Polyphen 2.0 [13] for predicting deleterious nsSNP. Imutant 3.0 [14] is a support vector-based method for protein stability prediction on single point mutations and Align-GVGD [15] examines the disease-associated nsSNPs/variants. For each method, protein sequence along with default parameters was submitted. A molecular dynamics studies was performed for evaluating the structural stability on the wild and predicted mutant proteins. Program package GROMACS 4.6.3 [16] with force field GROMOS96 [17] 43a1 using the Berendsen algorithm. Using XMGRACE graphs were plotted (Graphing, Advanced Computation and Exploration) program [18].
Results
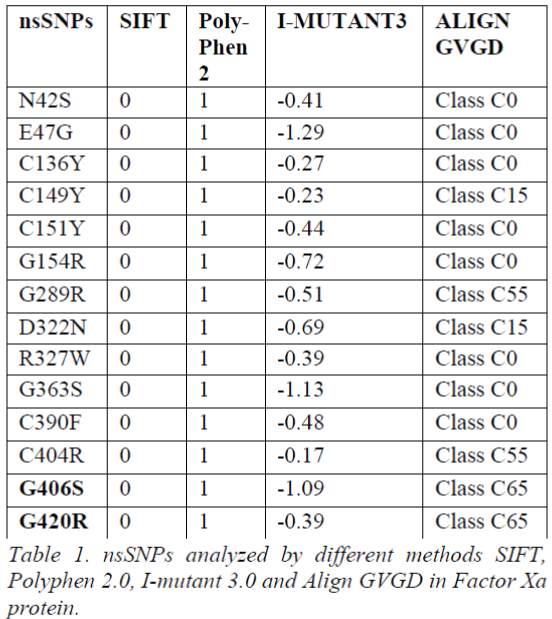
Tool Predictions
To determine the functional significance of the nsSNPs we utilized four most widely used computational tools in this analysis. Out of 58 investigated nsSNPs obtained from dbSNP and UniPort, 14 nsSNPs of Factor Xa gene were predicted to be deleterious by SIFT, Polyphen 2.0 and I-Mutant 3.0. Furthermore, we submitted 14 nsSNPs to Align GVGD. Based on the scores obtained from all the four methods, two nsSNPs G406S and G420R were predicted to be functional significant and deleterious causing disease (Table 1). To provide structural insight, an intensive molecular dynamics simulation was carried out for G406S and G420R. The molecular simulation may be useful to gain insight of nsSNPs on structural changes that affect the gene function.
Molecular Simulation Analysis
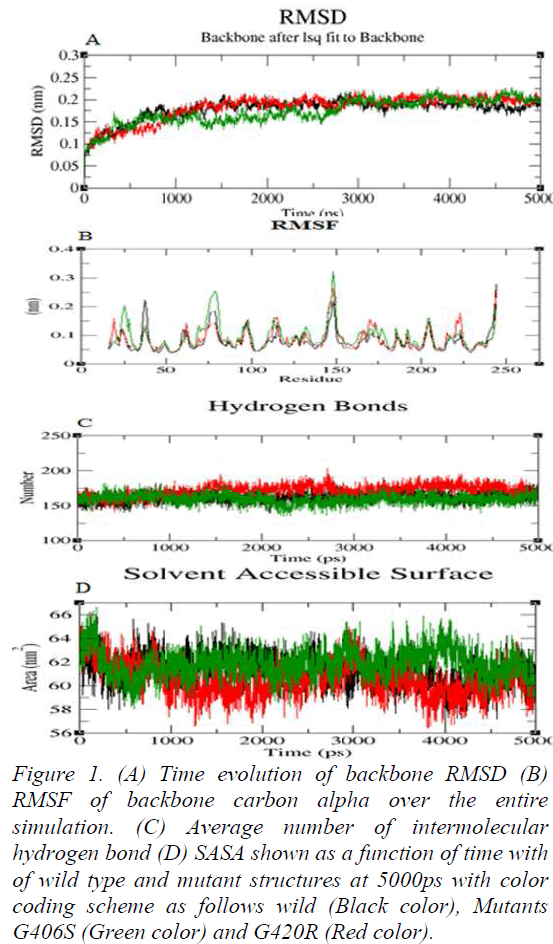
Molecular Dynamics simulation of wild and mutant structures were done with the aid of GROMACS 4.6.3; that adopts GROMOS96 43a1 force field parameter for energy minimizations. These structures of the wild 2VH6 and two mutant model were utilized to a base point that were solvated in a cubic box of 52.0Å nm divider and stacked with simple point charge water molecules. GENION method of GROMACS package was utilized to neutralize the structures by replacing irregular water atom with Na+ and Cl- particles. In order to neutralize the system, four Na+ molecules were added to replace four SPC water molecules. The systems investigated were subjected to steepest descent energy minimization reaching tolerances of 100kJ/mol. So as to soak the macromolecules into the water molecules, both the wild and mutantminimized structure were equilibrated for 50,000 steps at 300 K. After energy minimization, simulation was done in three steps: heating, equilibrating, and production. NVT (number of particles, volume, and temperature) was done at 300K and 1.0atm followed by NPT ensemble. At the final production simulation, the equilibrated structures of wild and mutant proteins were subjected to 50,000ps MD recreations. Using LINCS (Linear Constraint Solver) algorithm, all the covalent bonds were constrained [19]. The RMSD value of backbone atoms was calculated from trajectory file data. The nsSNP at 406 position from glycine to serine exhibited a deviation of ~0.20nm and glycine to arginine at 420 position exhibited a deviation of ~0.19nm from the wild protein of ~0.17nm at a range of 5000ps (Figure 1a). The deviation in wild and mutant structure display the mutation affected the dynamic behavior of the protein. From trajectory data generated for the wild and mutant proteins, RMSF values of the alpha carbon of each amino acid residue were analyzed. G406S and G420R showed the fluctuation of ~0.15nm and ~0.14nm with the wild protein of ~0.13nm respectively (Figure 1b). Notably, the rise in fluctuation was observed in mutant G406S. Hydrogen bonds play vital role in stability of the protein. Wild and mutant G406S showed ~162 hydrogen bonds whereas, mutant G420R showed more involvement of bonds by forming ~174 shown in (Figure 1c). The solvent accessibility surface area analysis predicted mutants G406S and G420R to be less exposed to surface area than wild. Wild type showed ~62nm whereas, mutant G406S showed ~61nm and ~60nm at 5000ps (Figure 1d). These results confirm that our prediction of deleterious nsSNPs at the position of G406S and G420R would play a critical role in causing disease.
Figure 1: (A) Time evolution of backbone RMSD (B) RMSF of backbone carbon alpha over the entire simulation. (C) Average number of intermolecular hydrogen bond (D) SASA shown as a function of time with of wild type and mutant structures at 5000ps with color coding scheme as follows wild (Black color), Mutants G406S (Green color) and G420R (Red color).
Discussion
When tissue injury occurs, a series of processes is involved to stop eventual blood loss. The process includes activation of extrinsic and intrinsic pathways of coagulation. Factor Xa is the site where two pathways converge which leads to the formation of insoluble fibrin at the injury site. An imbalance between clotting mechanism, clotting inactivation or thrombolytic processes can result in bleeding disorders. In this study, in silico analysis was employed to determine the deleterious nsSNPs in Factor Xa. Application of in silico prediction tools filter out variants of being deleterious and narrow down the search to specific variants for further experimental validation. These prediction algorithms have independent procedure in making their prediction which provides knowledge of mutational impact on protein structure and function. Furthermore, molecular dynamics simulation was used to study dynamic behaviour of wild and mutants types. In recent years many such studies has been carried out indicating the imapct fo mutation [23,24]. In order to examine the stability of the wild and mutant protein molecular dynamics simulations was initiated. In RMSD analysis G406S and G420R showed slight deviation from wild type. This might be due to physicochemical properties of the native and mutant amino acid. In G406S and G420R non-polar glycine is replaced by polar serine and polar basic arginine amino acid. In RMSF analysis mutant G406S showed higher fluctuation than wild protein. In an aspect of deviation, the mutant results in structural change with respect to flexibility compared to functional wild protein. In the stabilizing the protein structure, hydrogen bonds are the main driving force and play a vital role in biological interactions. Intramolecular hydrogen bond interaction is the most significant for protein stability. G406S showed less involvement in hydrogen formation when compared to G420R and wild type. Solvent accessibility area (SASA) accounts for the biomolecular surface area that is assessable to solvent molecules. Solvent accessible surface area for mutant G420R decreased comparatively to wild type where mutant G406S showed slight change through the simulation of 5000ps. The change in SASA values denotes the change in nature of the protein. Our obtained results are in concordance with experimental studies on the catalytic function of factor X [25]. Literature data suggest that subtitution of G406S cause defect in function of FX. We anticipate the obtained results from MD might provide new insights in disease diagnosis of Factor X at molecule level.
Conclusion
Molecular simulation performed on Factor Xa gene predicted mutants deviate from its wild-type leading to loss of stability and flexibility and resulting in diseaserelated mutations. These types of computational findings will help to establish the relationship between genetic variation and clinical phenotype to increase the current knowledge.
Acknowledgements
The authors take this opportunity to thank the management of VIT University for providing the facilities and encouragement to carry out this work.
References
- Mark Brooker. Registry of Clotting FactorConcentrates. World Federation of Hemophilia. 2008.
- Shaw KJ, Guilford WJ, Dallas JL, Koovakkaat SK, McCarrick MA, Liang A, Light DR, Morrissey MM. Journal of Medicinal Chemistry. 1998; 41: 3551.
- Fraternali F, Do QT, Doan BT, Atkinson RA, Palmas P, Sklenar V, Safar P, Wildgoose P, Strop P, Saudek V. Proteins Structure, Function, and Genetics.1998; 30: 264.
- Ewing WR, Becker MR, Manetta VE, Davis RS, Pauls HW, Mason H, Choi-Sledeski YM, Green D, Cha D, Spada AP, Cheney DL, Mason JS, MaignanS, Guilloteau JP, Brown K, Colussi D, Bentley R, Bostwick J, Kasiewski CJ, Morgan SR, Leadley RJ, Dunwiddie CT, Perrone MH, Chu VJ. Medicinal Chemistry. 1999; 42: 3557
- Rao MS, Olson AJ. Proteins: Structure, Function and Genetics. 1999; 34: 173.
- Böhm M, Stürzebecher J, Klebe G. Journal of Medicinal Chemistry. 1999; 42: 458.
- Vaz RJ, McLean LR, Pelton, JT, Journal Computer- Aided Molecular Design. 1998; 12-99.
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat T N, Weissig H, Shindyalov I N, & Bourne PE. The protein data bank.Nucleic Acids Research.2002, 28: 235-242.
- Kumar P, Henikoff S, & Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols.2009, 4: 1073-1082
- Adzhubei IA, Schmidt S, &Peshkin L. A method and server for predicting damaging missense mutations.Nature Methods.2010, 7: 248-249.
- Capriotti E, Fariselli P, Rossi I, &Casadio R. A three state prediction of single point mutations on protein stability changes. BMC Bioinformatics.2008, 9: article S6.
- Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A &Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. Journal of Medical Genetics, 2005, 43: 295-305.
- Hess B, Kutzner C, van der Spoel D, &Lindahl E. GROMACS 4: algorithms for highly efficient, loadbalanced, and scalable molecular simulation. Journal of Chemical Theory and Computation.2008, 4: 435-447.
- Jorgensen WL, Chandrasekhar J, Madura JD, ImpeyRW. Klein ML: Comparison of simple potential functions for simulating liquid water. Journal of Chemical Physics. 1983, 79-926.
- Turner PJ. XMGRACE, Version 5.1.19, Center For Coastal and Land-Margin Research, Oregon GraduGraduate Institute of Science and Technology, Beaverton, Ore. 2005.
- Hess B, Bekker H, Berendsen HJ, Fraaije JGEM. LINCS:alinearconstraint solver for molecular simulations. Journal of Computational Chemistry.1997, 18: 1463–1472.
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle meshEwald method. Journal of Chemical Physics.1995, 103: 8577–8593.
- George PDC. Computational Methods in SNP Analysis.Encyclopedia of Life Sciences. 2014, DOI: 10.1002/9780470015902.a0025492.
- George PDC &Sethumadhavan R. Investigation onthe role of nsSNPs in HNPCC genes-a bioinformatics approach. Journal of Biomedical Science.2009, 16:42.
- Isshiki I, Favier R, Moriki T, Uchida T, Ishihara H, Van Dreden P, Murata M &Ikeda Y. Genetic analysis of hereditary factor X deficiency in a French patient of Sri Lankan ancestry: in vitro expression study identified Gly366Ser substitution as the molecular basis of the dysfunctional factor X. Journal of blood coagulation and Fibrinolysis. 2005, 16: 9- 16.
- Abdel AS, Oliva R, Chermak E, De Cristofaro R, Cavallo L. Molecular dynamics characterization of five pathogenic factor X mutants associated with decreased catalytic activity. Journal of Biochemistry.2014, 53: 44.
- Vianello F, Lombardi AM, Boldrin C, Luni S, GirolamiA. A new factor X defect (factor X padua 3) : a compound heterozygous between true deficiency ( Gly(380)-Arg) and an abnormality (Ser(334)-Pro). Journal of Thrombosis Research. 2001, 104: 257- 264.