ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Rapid Communication - Biomedical Research (2018) Volume 29, Issue 14
Exosomes released from pancreatic cancer cells are heterogeneous particle populations
1Department of Bioscience and Laboratory Medicine, Graduate School of Health Sciences, Hirosaki University, 66-1 Hon-Cho, Hirosaki, Aomori, Japan
2Department of Radiation Sciences, Graduate School of Health Sciences, Hirosaki University, 66-1 Hon-Cho, Hirosaki, Aomori, Japan
- *Corresponding Author:
- Mitsuru Chiba
Department of Bioscience and Laboratory Medicine
Graduate School of Health Sciences
Hirosaki University
66-1 Hon-Cho, Hirosaki, Aomori, Japan
Accepted date: July 16, 2018
DOI: 10.4066/biomedicalresearch.29-18-782
Visit for more related articles at Biomedical ResearchExosomes are extracellular vesicles that are released from cells and comprise microRNAs (miRNAs); however, little is known about the homogeneity of their contents. In this study, we assessed the homogeneity of exosomes by measuring the number of particles and miRNA copies released by pancreatic cancer PK-45H cells. miR-204-3p and miR-638 were abundant in the exosomes released by PK-45H cells and were present as one-to-several copies per 10,000 particles. The number of miR-638 copies released by PK-45H cells significantly declined following treatment with 10 μM GW4869, a neutral sphingomyelinase 2 (nSMase2) inhibitor. These findings suggest that exosomes form heterogeneous particle populations and nSMase2 may be involved in the exosomal uptake of miR-638.
Keywords
Exosomes, Extracellular vesicles, MicroRNA, Pancreatic cancer cell, GW4869, Heterogeneity.
Introduction
Exosomes are extracellular vesicles (EVs) that are approximately 40-200 nm in diameter and are released by several cells. In 1983, Pan et al. reported the presence of exosomes as vesicles secreted by sheep reticulocytes [1]. Exosomes comprise a lipid bilayer and tetraspanin proteins on their membrane surfaces [2]. Among the tetraspanin protein family, CD63, CD9, and CD81 are known to be frequently localized at the exosomal surfaces and are therefore used as exosomal markers [2]. In addition, exosomes contain cytoskeletal proteins, Rab family proteins, heat shock proteins and enzymes, as well as proteins involved in signal transduction [2]. Valadi et al. reported that exosomes containing mRNAs and microRNAs (miRNAs) can be delivered to other cells [3]. Exosomal proteins and RNAs that have been reported are described in the ExoCarta database (http://exocarta.ludwig.edu.au) [4]. It has also been reported that neutral sphingomyelinase 2 (nSMase2) is involved in the secretion of exosomal miRNAs [5,6] and that GW4869, which is a known nSMase2 inhibitor, inhibits exosome secretion [5]; however, its effects on miRNA copy numbers remain unclear.
Observations made using electron microscopy suggest that exosomes may represent heterogeneous particle populations; however, the heterogeneity of exosomal components released by specific cells is unknown. In this study, we assessed the heterogeneity of exosomes by particle measurement using miRNA copies released from pancreatic cancer PK-45H cells.
In addition, we examined the changes in the particle counts and miRNA copies released into the culture supernatants by PK-45H cells treated with GW4869.
Materials and Methods
Cell line
The human pancreatic cancer cell line PK-45H was purchased from Riken BRC (Ibaraki, Japan). PK-45H cells were cultured using RPMI1640 medium (Wako, Tokyo, Japan) supplemented with 10% fetal bovine serum (ThermoFisher Scientific, Carlsbad, CA, USA), 100 U/ml of penicillin, and 100 μg/ml of streptomycin (Wako) at 37°C in a humidified atmosphere of 5% CO2.
Purification of exosomes
PK-45H cells were plated on 10-cm dishes at a density of 2 × 105 cells per dish in the culture medium. This medium was discarded after 4 d, the dishes were washed thrice with serumfree RPMI1640 medium, and 10 ml of serum-free culture medium with 0, 1.0, 3.3, or 10 μM of GW4869 (Sigma- Aldrich, St. Louis, MO, USA), a nSMase2 inhibitor, was added to each dish. Cell culture media were collected after 48 h. Exosomes from the media were differentially centrifuged and purified using a previously reported procedure [7]. Exosomal pellets were suspended in Dulbecco’s phosphate buffered saline (D-PBS (-)) and the exosomal particles were measured using a NanoSight LM10 particle analyzer (NanoSight, Malvern, UK).
RNA extraction
Exosomal RNAs were isolated using Isogen II reagent (NipponGene, Tokyo, Japan) according to the manufacturer’s instructions.
Human miRNA microarrays
Exosomal miRNAs were labeled using a miRCURY LNA miRNA Hy5 Power labeling kit (Exiqon, Copenhagen, Denmark). Hy5-labeled miRNAs in a hybridization solution were incubated on a Toray 3D-Gene human miRNA oligo chip (Toray, Tokyo, Japan) at 32°C for 16 h using a hybridization oven. Next, hybridized microarray chips were washed in a wash buffer according to the manufacturer’s instructions, and the images of fluorescent signals were captured using a Toray 3D-Gene scanner 3000 (Toray).
Real-time PCRcDNAs of miR-204-3p and miR-638 oligo RNA were diluted to produce standard curves, whereas miR-204-3p and miR-638 in the culture supernatants were reverse transcribed using TaqMan microRNA RT kits and the appropriate 5 × RT primers (both from Thermo Fisher Scientific) according to the manufacturer’s instructions. Subsequently, qPCR was performed using a FastStart TaqMan probe master kit (Roche Diagnostics, Basel, Switzerland), 20 × of each probe, and a StepOne Plus real-time PCR system (Thermo Fisher Scientific) at the following conditions: 10 min at 95°C, followed by 40 cycles at 95°C for 15 s and 60°C for 60 s. Copy numbers of each miRNA were calculated using the standard curve.
Statistical analysis
Statistical significance (p<0.05) was determined by the Tukey- Kramer test for multiple comparisons using Statcell 3 software (OMS publishing Inc., Saitama, Japan).
Results and Discussion
Exosomes have been suggested to form a heterogeneous population based on electron microscopic observations; however, the heterogeneity of exosome particles released from specific cells has not been demonstrated. In this study, we evaluated the heterogeneity of the exosome particles released from the PK-45H pancreatic cancer cells by measuring the particle numbers in supernatants and by quantifying the miRNA copy numbers.
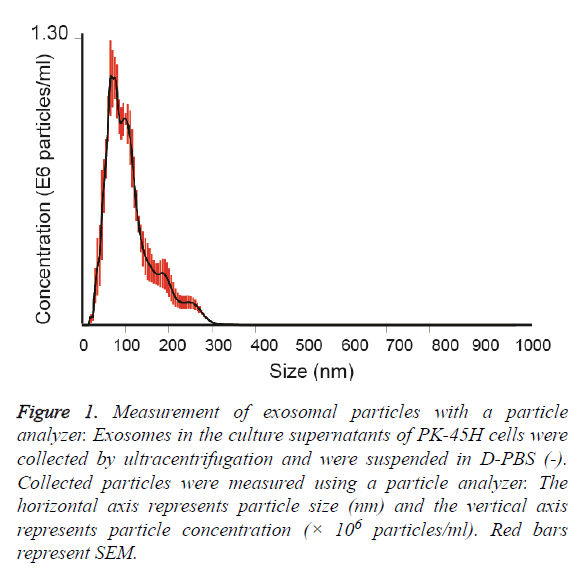
Exosomes released by PK-45H cell were collected by ultracentrifugation and the particle sizes of the vesicles were measured with a particle analyzer. The average particle size was 111 ± 7.4 nm (Figure 1). Other studies have also reported the particle sizes of such vesicles at approximately 100 nm [8,9], indicating that the vesicles collected in this study revealed the characteristics of exosomes.
Figure 1: Measurement of exosomal particles with a particle analyzer. Exosomes in the culture supernatants of PK-45H cells were collected by ultracentrifugation and were suspended in D-PBS (-). Collected particles were measured using a particle analyzer. The horizontal axis represents particle size (nm) and the vertical axis represents particle concentration (× 106 particles/ml). Red bars represent SEM.
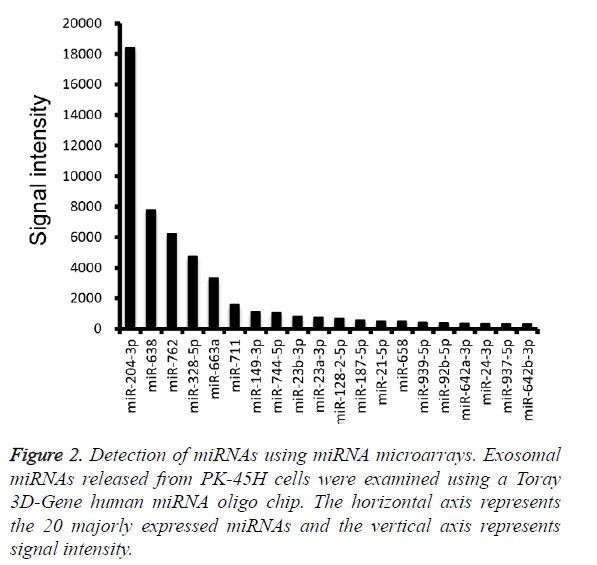
Furthermore, miRNA microarray analysis was performed to identify the miRNA species in the PK-45H-derived exosomes. The 20 most abundant miRNAs are presented in Figure 2, and among these, we focused on miR-204-3p and miR-638.
Fibronectin 1, a target gene of miR-204-3p, suppresses the proliferation of tumor vascular endothelial cells of liver cancer [10], whereas cyclin-dependent kinase 2 (CDK2), SPY-box2 (SOX2), and specificity protein 2 (SP2) are the recognized target genes of miR-638 [11-13].
CDK2 inhibits differentiation and proliferation of leukemia cells [11], SOX2 suppresses invasion of colorectal cancer cells and SP2 suppresses cell proliferation of several digestive organ cancers [12,13]. These observations indicate that miR-204-3p and miR-638 are tumor-suppressing miRNAs and suggest that PK-45H cells may maintain homeostasis and function as cancer cells by expelling those miRNAs into the extracellular space.
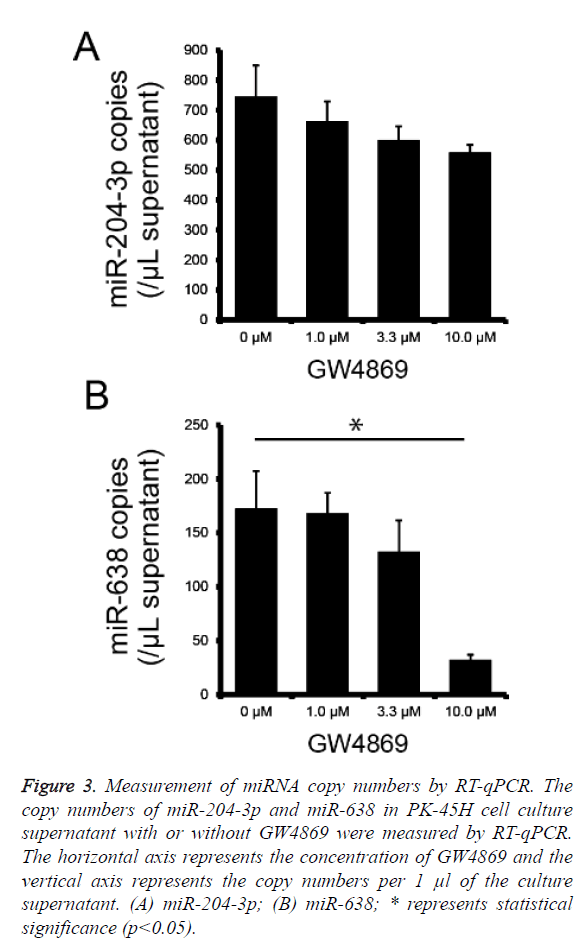
To determine whether miR-204-3p and miR-638 are secreted by PK-45H cells via the nSMase2 pathway, we performed absolute quantification of miR-204-3p and miR-638 copy numbers using real-time PCR on the culture supernatants of PK-45H cells treated with 0, 1.0, 3.3, or 10 μM of GW4869 and calculated miR-204-3p and miR-638 copy numbers per 1 μl of supernatant. The secretion of miR-204-3p decreased with the increasing GW4869 concentration; however, this was not statistically significant (Figure 3a), whereas the secretion of miR-638 significantly decreased at 10 μM of GW4869 (Figure 3b). These findings suggest that nSMase2 is crucial in the extracellular secretion of miR-638. We previously reported that miR-638 secretion in the human hepatocellular cancer HuH-7 cells is also inhibited by treatment with GW4869 [14]. This new result is consistent with our previous work, suggesting that miR-638 is released from at least the pancreatic and hepatocellular cancer cells via the nSMase2 pathway.
Figure 3: Measurement of miRNA copy numbers by RT-qPCR. The copy numbers of miR-204-3p and miR-638 in PK-45H cell culture supernatant with or without GW4869 were measured by RT-qPCR. The horizontal axis represents the concentration of GW4869 and the vertical axis represents the copy numbers per 1 μl of the culture supernatant. (A) miR-204-3p; (B) miR-638; * represents statistical significance (p<0.05).
To assess the homogeneity of particles using the exosome counts and miR-204-3p and miR-638 copy numbers, we calculated the copy numbers per particle and examined the effects of GW4869 treatment. The particle counts did not change following treatment with GW4869 (Table 1), indicating that inhibition of the nSMase2 pathway does not influence particle release. miR-204-3p and miR-638 were present as oneto- several copies per 10,000 particles and miR-638 copy number decreased by treatment with 10 μM of GW4869 (Table 1). This suggests that nSMase2 is not involved in the particle formation but rather in the uptake of miR-638 into exosomes.
| GW4869 | ||||
|---|---|---|---|---|
| 0 µM | 1.0 µM | 3.3 µM | 10.0 µM | |
| EV counts (× 109 EVs/mL) | 1.24 ± 0.17 | 1.07 ± 0.07 | 0.95 ± 0.03 | 1.09 ± 0.10 |
| miR-204-3p copies (/µL supernatant) | 746.38 ± 229.19 | 662.91 ± 147.89 | 600.09 ± 102.01 | 559.60 ± 55.02 |
| miR-638 copies (/µL supernatant) | 172.60 ± 77.71 | 168.31 ± 41.60 | 132.60 ± 64.48 | 32.33 ± 10.25 |
| miR-204-3p copies (/10000 EVs) | 6.02 ± 1.85 | 6.20 ± 1.38 | 6.32 ± 1.07 | 5.13 ± 0.50 |
| miR-638 copies (/10000 EVs) | 1.39 ± 0.63 | 1.57 ± 0.39 | 1.40 ± 0.68 | 0.30 ± 0.09 |
Table 1: The copy numbers of miR-204-3p and miR-638 into EVs released from PK-45H cells treated with GW4869.
Pancreatic cancer is one of the deadliest cancers with rapid disease progression, requiring further elucidation of its underlying molecular mechanisms and novel biomarkers for early detection [15]. Pancreatic cancer cells intimately interact with a complex microenvironment that influences pancreatic cancer progression.
Exosomes are emerging as important mediators of the crosstalk between tumor cells and the microenvironment of pancreatic cancer [16]. Masamune et al. reported that exosome-like EVs released from pancreatic cancer cells induced the AKT phosphorylation and mitogen-activated protein kinase signaling in pancreatic stellate cells [17]. However, the effects of exosomal miR-204-3p and miR-638 into other pancreatic cancer cells or stromal cells remain unknown. In future, we will examine whether exosomal miR-204-3p and miR-638 is involved in pancreatic cancer progression or not. In addition, studying the separation of exosome particles including miRNAs should be considered.
Competing Interest
All authors declare that they have no conflict of interests or financial interests.
Acknowledgement
This work was supported in part by a Hirosaki University Institutional Research Grant for Young Scientists, and JSPS KAKENHI (no. 17H04761 and 17K19779).
References
- Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 1983; 33: 967-978.
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 2010; 73: 1907-1920.
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9: 654-659.
- Simpson RJ, Kalra H, Mathivanan S. ExoCarta as a resource for exosomal research. J Extracell Vesicles 2012; 1: 18374.
- Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 2010; 285: 17442-17452.
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008; 319: 1244-1247.
- Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep 2012; 28: 1551-1558.
- Webber J, Clayton A. How pure are your vesicles? J Extracell Vesicles 2013; 2: 19861.
- Lang FM, Hossain A, Gumin J, Momin EN, Shimizu Y, Ledbetter D, Shahar T, Yamashita S, Parker Kerrigan B, Fueyo J, Sawaya R, Lang FF. Mesenchymal stem cells as natural biofactories for exosomes carrying miR-124a in the treatment of gliomas. Neuro Oncol 2018; 20: 380-390.
- Cui ZH, Shen SQ, Chen ZB, Hu C. Growth inhibition of hepatocellular carcinoma tumor endothelial cells by miR-204-3p and underlying mechanism. World J Gastroenterol 2014; 20: 5493-5504.
- Lin Y, Li D, Liang Q, Liu S, Zuo X, Li L, Sun X, Li W, Guo M, Huang Z. miR-638 regulates differentiation and proliferation in leukemic cells by targeting cyclin-dependent kinase 2. J Biol Chem 2015; 290: 1818-1828.
- Ma K, Pan X, Fan P, He Y, Gu J, Wang W, Zhang T, Li Z, Luo X. Loss of miR-638 in vitro promotes cell invasion and a mesenchymal-like transition by influencing SOX2 expression in colorectal carcinoma cells. Mol Cancer 2014; 13: 118.
- Zhao LY, Yao Y, Han J, Yang J, Wang XF, Tong DD, Song TS, Huang C, Shao Y. miR-638 suppresses cell proliferation in gastric cancer by targeting Sp2. Dig Dis Sci 2014; 59: 1743-1753.
- Kubota S, Chiba M, Watanabe M, Sakamoto M, Watanabe N. Secretion of small/microRNAs including miR-638 into extracellular spaces by sphingomyelin phosphodiesterase 3. Oncol Rep 2015; 33: 67-73.
- Siegel RL, Miller KD and Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68: 7-30.
- Nazarenko I, Rana S, Baumann A, Nazarenko I, Rana S, Baumann A, McAlear J, Hellwig A, Trendelenburg M and Lochnit G. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res 2010; 70: 1668-1678.
- Masamune A, Yoshida N, Hamada S, Takikawa T, Nabeshima T and Shimosegawa T. Exosomes derived from pancreatic cancer cells induce activation and profibrogenic activities in pancreatic stellate cells. Biochem Biophys Res Commun 2018; 495: 71-77.