ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2019) Volume 30, Issue 2
Bioinformatical analysis of gene expressions and pathways in human colorectal cancer tissues
Department of Bioscience and Laboratory Medicine, Graduate School of Health Sciences, Hirosaki University, Japan
- *Corresponding Author:
- Mitsuru Chiba
Department of Bioscience and Laboratory Medicine
Graduate School of Health Sciences, Hirosaki University
66-1 Hon-cho, Hirosaki, Aomori 036-8564, Japan
Accepted Date: January 19, 2019
DOI: 10.35841/biomedicalresearch.30-19-022
Visit for more related articles at Biomedical ResearchIntroduction: Colorectal cancer (CRC) is a major disease and a leading cause of mortality and morbidity worldwide. The expression of various genes changes in CRC. This study aimed to investigate differentially expressed genes (DEGs) in CRC tissues and to predict important pathways associated with CRC. Methods: DEGs between cancerous and non-cancerous tissues from 17 patients with CRC were screened using Affymetrix HG-U133 Plus 2.0 microarray data downloaded from the Gene Expression Omnibus (GSE32323). Next, functional and pathway analysis of DEGs was performed by Gene Ontology (GO) analysis and WikiPathways analysis using GeneSpring 14.5 software. Results and discussion: Compared with non-cancerous tissues, 3,856 DEGs were identified in the CRC samples, which included 2,015 upregulated DEGs and 1,841 downregulated DEGs, with the selection criteria of p < 0.05 and a two-fold change. The GO analysis identified 448 and 606 GO terms using the upregulated and downregulated DEGs, respectively. Interestingly, exosomes and extracellular vesicles were associated with the downregulated DEGs. A total of 240 pathways were associated with DEGs in CRC tissues. Taken above, I have identified candidate DEGs in CRC tissues and pathways. These candidate genes and pathways could be therapeutic targets for CRC.
Keywords
Bioinformatics, Colorectal cancer, Gene expression, Pathway, Gene ontology.
Introduction
Colorectal cancer (CRC) is the fourth most common type of cancer in the United States and the second leading cause of death due to cancer [1]. Recently, the incidence and mortality rates of colorectal cancer have increased in Japan. It is estimated that about 10% of men and 8% of women in Japan will be diagnosed with CRC during their lifetime [2]. CRC affects all racial and ethnic groups and is the most common in people aged 50 years and older. The stage of the cancer is indicative of how far it has spread, and determining the stage helps in choosing the most appropriate treatment. However, CRC remains a prominent global health problem, which may be attributed to a lack of comprehensive and systemic understanding of the underlying molecular mechanisms of carcinogenesis and progression.
CRC was one of the first solid tumours that was molecularly characterized, with several genes and pathways implicated in tumour initiation and growth [3]. CRC carcinogenesis is the result of a stepwise accumulation of genetic events occurring in oncogenes and tumour suppressor genes that deregulate the key signaling pathways that drive progression, such as Wnt/β- catenin [4], transforming growth factor beta (TGF-β) [5], epidermal growth factor receptor (EGFR) [6], mitogenactivated protein kinase and phosphoinositide 3-kinase (PI3K)-Akt pathways [7,8]. Therefore, CRC carcinogenesis and progression involve the activation or suppression of a complex network of molecules. However, the understanding of CRC carcinogenesis and progression networks is insufficient.
Khamas et al. examined gene expression profiles of cancerous and non-cancerous tissues from 17 patients with CRC using Affimetrix GeneChip Human Genome U133 Plus 2.0 Array [9]. In the present study, a bioinformatical analysis was performed to identify genes and pathways involved in CRC using these data sets (GSE32323).
Materials and Methods
Data source
The gene expression profile of GSE32323 was obtained from the Gene Expression Omnibus database. This dataset contains 17 pairs of cancer and non-cancerous tissues from CRC patients [9]. Microarray analysis was performed using the Affimetrix GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix Inc., Santa Clara, CA, USA).
Data processing and screening for differentially expressed genes (DEGs)
Further, microarray data were normalized using GeneSpring 14.5 software (Agilent Technologies, Foster City, CA, USA). MAS5 was used as a summarization algorithm. Baseline transformation was not performed. A lower cut-off value of 10 was used to filter entities in which at least 17 out of 34 samples had values within the range were retained. Only acceptable flags "Present" were used. I retained entities in which at least 17 out of 34 samples have acceptable values.
The threshold criterion for the selection of DEGs was fold change of >2.0. Statistical analysis was performed using a moderated t-test with P<0.05. Benjamini Hochberg False Discovery Rate procedure was used for controlling falsepositives in multiple testing corrections. Selected DEGs were presented using volcano and scatter plots.
Functional enrichment analysis of DEGs
To examine functions and pathways that may be altered by the upregulated or downregulated DEGs, Gene Ontology (GO) analysis and WikiPathways analysis was performed using GeneSpring 14.5 software (Agilent Technologies). Pathway data of Homo sapiens were downloaded from WikiPathways. The cut-off value for the screening of significant functions and pathways was P<0.05.
Result and Discussion
Identification of DEGs between CRC and normal samples
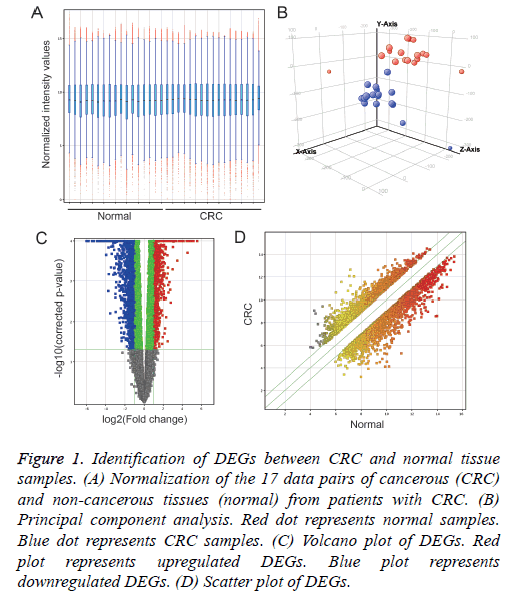
Normalization of each 17 pair data of cancer (CRC) and noncancerous tissues (normal) from CRC patients (Figure 1A) and Principal component analysis (Figure 1B) was performed using GeneSpring 14.5 software. Compared with normal samples, 3,856 DEGs were identified in CRC samples, including 2,015 up-regulated DEGs and 1,841 down-regulated DEGs (Figures 1C and 1D).
Figure 1: Identification of DEGs between CRC and normal tissue samples. (A) Normalization of the 17 data pairs of cancerous (CRC) and non-cancerous tissues (normal) from patients with CRC. (B) Principal component analysis. Red dot represents normal samples. Blue dot represents CRC samples. (C) Volcano plot of DEGs. Red plot represents upregulated DEGs. Blue plot represents downregulated DEGs. (D) Scatter plot of DEGs.
Analysis of biological functions and pathways altered by DEGs
Under p-value of 0.05, 448 GO terms were identified in the upregulated DEGs, and 606 GO terms were identified in the downregulated DEGs. Of these, the top 15 GO terms are presented in Table 1. Notably, cell-to-cell communication tool such as extracellular exosome (P=1.11 × 10-28) or extracellular vesicle (P=2.67 × 10-28) was significantly associated with the downregulated DEGs (Table 1). Exosomes are a subclass of extracellular vesicles that are released by all cell types, including cancer cells, and are involved in intercellular communication [10]. These exosomes have been detected in the cancer microenvironment, and emerging evidence suggests that they play a role in facilitating tumorigenesis by regulating angiogenesis, immunity, and metastasis [11]. This indicates that cell-to-cell communication is abnormal in the CRC microenvironment. Thus, analysis of exosomes is necessary for a better understanding of CRC carcinogenesis and progression in future studies.
| GO number | GO term | Count | P-value |
|---|---|---|---|
| Up-Regulation | |||
| GO:0070013 | Intracellular organelle lumen | 652 | 0 |
| GO:0034641 | Cellular nitrogen compound metabolic process | 672 | 0 |
| GO:0006139|GO:0055134 | Nucleobase-containing compound metabolic process | 603 | 0 |
| GO:0031974 | Membrane-enclosed lumen | 652 | 0 |
| GO:0031981 | Nuclear lumen | 580 | 0 |
| GO:0043233 | Organelle lumen | 652 | 0 |
| GO:0044428 | Nuclear part | 620 | 0 |
| GO:0005634 | Nucleus | 847 | 0 |
| GO:0046483 | Heterocycle metabolic process | 618 | 1.40 x 10-45 |
| GO:0006725 | Cellular aromatic compound metabolic process | 622 | 1.40 x 10-45 |
| GO:0090304 | Nucleic acid metabolic process | 552 | 2.80 x 10-45 |
| GO:0000278|GO:0007067 | Mitotic cell cycle | 196 | 4.20 x 10-45 |
| GO:0005654 | Nucleoplasm | 464 | 4.20 x 10-45 |
| GO:1901360 | Oganic cyclic compound metabolic process | 634 | 3.01 x 10-43 |
| GO:0006807 | Nitrogen compound metabolic process | 915 | 9.17 x 10-38 |
| Down-Regulation | |||
| GO:0043230 | Extracellular organelle | 367 | 2.86 x 10-28 |
| GO:0070062 | Extracellular exosome | 367 | 1.11 x 10-28 |
| GO:1903561 | Extracellular vesicle | 367 | 2.67 x 10-28 |
| GO:0071944 | Cell periphery | 530 | 1.14 x 10-26 |
| GO:0044421 | Extracellular region part | 454 | 3.99 x 10-26 |
| GO:0005615 | Extracellular space | 435 | 6.98 x 10-26 |
| GO:0005886|GO:0005904 | Plasma membrane | 516 | 1.61 x 10-25 |
| GO:0031982|GO:0031988 | Vesicle | 459 | 1.31 x 10-23 |
| GO:0005576 | Extracellular region | 485 | 1.53 x 10-20 |
| GO:0044425 | Membrane part | 641 | 4.48 x 10-19 |
| GO:0070887 | Cellular response to chemical stimulus | 315 | 1.09 x 10-18 |
| GO:0016020 | Membrane | 808 | 1.31 x 10-17 |
| GO:0065008 | Regulation of biological quality | 406 | 1.52 x 10-17 |
| GO:0006629 | Lipid metabolic process | 166 | 3.25 x 10-17 |
| GO:0050896|GO:0051869 | Response to stimulus | 718 | 4.37 x 10-17 |
Table 1: Gene Ontology (GO) analysis and significant top 15 GO terms of DEGs in human colorectal cancer.
Current CRC screening methods are divided into invasive and non-invasive tests. The non-invasive tests include stool and blood-based tests (fecal immunochemical test and fecal DNA test etc.) and radiologic tests (computed tomographic colonography etc.) [12]. Recently, exosomal components of blood samples have been studied for CRC biomarkers [13]. The ideal screening study should be efficient with high sensitivity and specificity, safe, available, convenient, and cheap.
Using a cut-off p-value of 0.05, 240 pathways were detected that involved up-regulated and down-regulated DEGs. Of these, the top 30 pathways are presented in Table 2. Using one of these pathways, the relationship among TGF-β, PI3K-Akt, and vascular endothelial growth factor (VEGF)-A-VEGF receptor 2 (VEGFR2) signaling pathways was predicted. The TGF-β signaling pathway plays an important role in cancer cell proliferation, growth, metastasis, and apoptosis and elicits both pro- and anti-oncogenic effects [5]. This pathway is controlled by positive regulators such as Smad2, Smad3, and Smad4 and negative regulators such as Smad6 and Smad7. Mutations in the TGF-β signaling components, including TGF-β receptors and cytoplasmic signaling transducers, are frequently observed in CRC. The regulatory effects of the TGF-β/Smad signaling pathway on cell growth inhibition and apoptosis contribute to CRC progression. Further, Smad4 deletion is a poor response marker in patients undergoing chemotherapy, whereas Smad7 deletion is a good prognostic marker in patients with CRC.
| Pathway name | P-value | Gene count |
|---|---|---|
| Hs_Glucuronidation_WP698_94183 | 6.32 x 10-11 | 26 |
| Hs_miRNA_Regulation_of_DNA_Damage_Response_WP1530_98336 | 1.83 x 10-10 | 98 |
| Hs_G1_to_S_cell_cycle_control_WP45_97000 | 1.94 x 10-10 | 65 |
| Hs_DNA_Replication_WP466_96302 | 2.10 x 10-10 | 42 |
| Hs_DNA_Damage_Response_WP707_94731 | 2.11 x 10-10 | 68 |
| Hs_Hepatitis_C_and_Hepatocellular_Carcinoma_WP3646_102150 | 2.55 x 10-10 | 57 |
| Hs_DNA_IR-damage_and_cellular_response_via_ATR_WP4016_101923 | 2.66 x 10-10 | 83 |
| Hs_Retinoblastoma_Gene_in_Cancer_WP2446_98748 | 2.99 x 10-10 | 90 |
| Hs_Cell_Cycle_WP179_97174 | 3.53 x 10-10 | 120 |
| Hs_TGF-beta_Signaling_Pathway_WP366_90028 | 3.78 x 10-10 | 132 |
| Hs_Focal_Adhesion-PI3K-Akt-mTOR-signaling_pathway_WP3932_102154 | 6.36 x 10-10 | 302 |
| Hs_PI3K-Akt_Signaling_Pathway_WP4172_96453 | 6.54 x 10-10 | 340 |
| Hs_Nuclear_Receptors_Meta-Pathway_WP2882_97489 | 6.57 x 10-10 | 318 |
| Hs_VEGFA-VEGFR2_Signaling_Pathway_WP3888_102004 | 9.53 x 10-10 | 236 |
| Hs_Myometrial_Relaxation_and_Contraction_Pathways_WP289_96894 | 1.07 x 10-9 | 156 |
| Hs_Prostaglandin_Synthesis_and_Regulation_WP98_98180 | 1.24 x 10-9 | 46 |
| Hs_Metapathway_biotransformation_Phase_I_and_II_WP702_102200 | 1.86 x 10-9 | 190 |
| Hs_Gastric_Cancer_Network_1_WP2361_101906 | 2.17 x 10-9 | 29 |
| Hs_Imatinib_and_Chronic_Myeloid_Leukemia_WP3640_89384 | 4.20 x 10-9 | 21 |
| Hs_Adipogenesis_WP236_97606 | 5.13 x 10-9 | 131 |
| Hs_Vitamin_D_Receptor_Pathway_WP2877_94793 | 5.38 x 10-9 | 186 |
| Hs_Breast_cancer_pathway_WP4262_97132 | 6.51 x 10-9 | 156 |
| Hs_NRF2_pathway_WP2884_94787 | 8.53 x 10-9 | 143 |
| Hs_LncRNA_involvement_in_canonical_Wnt_signaling_and_colorectal_cancer_WP4258_97136 | 8.70 x 10-9 | 98 |
| Hs_Integrated_Cancer_Pathway_WP1971_98351 | 8.87 x 10-9 | 49 |
| Hs_Regulation_of_sister_chromatid_separation_at_the_metaphase-anaphase_transition_WP4240_96623 | 1.05 x 10-8 | 15 |
| Hs_Focal_Adhesion_WP306_97459 | 1.84 x 10-8 | 198 |
| Hs_Circadian_rythm_related_genes_WP3594_87161 | 2.43 x 10-8 | 210 |
| Hs_Pyrimidine_metabolism_WP4022_95459 | 2.84 x 10-8 | 99 |
| Hs_ESC_Pluripotency_Pathways_WP3931_102153 | 5.31 x 10-8 | 116 |
Table 2: Top 30 of 240 pathways enriched with DEGs in CRC.
The PI3K–Akt signaling pathway leads to reduced apoptosis, stimulates cell growth and increases proliferation [8]. Genetic aberrations leading to PI3K-Akt hyper-activation are observed at considerable frequency in all major nodes in most tumors. In CRC the most commonly observed pathway changes are insulin-like growth factor 2 overexpression, phosphatidylinositol-4,5-bisphosphate-3- kinase catalytic subunit alpha mutations and phosphatase and tensin homolog deleted from chromosome 10 mutations and deletions. VEGFR2 signal transduction and trafficking pathways are mediated by VEGF-A ligands [14]. VEGFAVEGFR2 signaling pathway is involved in cell proliferation, chemotaxis and survival of endothelial cells. Angiogenesis is mainly regulated by this signaling pathway. In CRC, this pathway is associated with survival and distant metastasis of cancer cells.
The above mentioned pathway analyses suggested that the microarray data obtained in this study was indicative for colon cancer. As a novel pathway, the involvement of lncRNA in canonical Wnt signaling in CRC was predicted (Table 2). Taken above, novel candidate genes and pathways in CRC were identified by integrated bioinformatical analysis. These candidate genes and pathways may act as potential therapeutic targets for CRC.
Conclusion
I have identified candidate DEGs in CRC tissues and pathways. These candidate genes and pathways could be therapeutic targets for CRC.
Acknowledgement
This work was supported in part by a Hirosaki University Institutional Research Grant for Young Scientists, JSPS KAKENHI (no. 23790613), a Grant-in-Aid for Young Scientists (B), and a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT).
References
- Wolf AMD, Fontham ETH, Church TR, Flowers CR, Guerra CE, LaMonte SJ, Etzioni R, McKenna MT, Oeffinger KC, Shih YT, Walter LC, Andrews KS, Brawley OW, Brooks D, Fedewa SA, Manassaram-Baptiste D, Siegel RL, Wender RC, Smith RA. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin 2018; 68: 250-281.
- Tamakoshi A, Nakamura K, Ukawa S, Okada E, Hirata M, Nagai A, Matsuda K, Kamatani Y, Muto K, Kiyohara Y, Yamagata Z, Ninomiya T, Kubo M, Nakamura Y; BioBank Japan Cooperative Hospital Group. Characteristics and prognosis of Japanese colorectal cancer patients: The BioBank Japan Project. J Epidemiol 2017; 27: S36-S42.
- Dienstmann R, Vermeulen L, Guinney J, Kopetz S, Tejpar S, Tabernero J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer 2017; 17: 79-92.
- Pandurangan AK, Dharmalingam P, Sadagopan SK, Ramar M, Munusamy A, Ganapasam S. Luteolin induces growth arrest in colon cancer cells through involvement of Wnt/Ã-catenin/GSK-3Ã signaling. J Environ Pathol Toxicol Oncol 2013; 32: 131-139.
- Soleimani A, Pashirzad M, Avan A, Ferns GA, Khazaei M, Hassanian SM. Role of the transforming growth factor-Ã signaling pathway in the pathogenesis of colorectal cancer. J Cell Biochem 2018.
- Bertotti A, Papp E, Jones S, Adleff V, Anagnostou V, Lupo B, Sausen M, Phallen J, Hruban CA, Tokheim C, Niknafs N, Nesselbush M, Lytle K, Sassi F, Cottino F, Migliardi G, Zanella ER, Ribero D, Russolillo N, Mellano A, Muratore A, Paraluppi G, Salizzoni M, Marsoni S, Kragh M, Lantto J, Cassingena A, Li QK, Karchin R, Scharpf R, Sartore-Bianchi A, Siena S, Diaz LA Jr, Trusolino L, Velculescu VE. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature 2015; 526: 263-267.
- Zhao J, Ou B, Han D, Wang P, Zong Y, Zhu C, Liu D, Zheng M, Sun J, Feng H, Lu A. Tumor-derived CXCL5 promotes human colorectal cancer metastasis through activation of the ERK/Elk-1/Snail and AKT/GSK3Ã/Ã-catenin pathways. Mol Cancer 2017; 16: 70.
- Danielsen SA, Eide PW, Nesbakken A, Guren T, Leithe E, Lothe RA. Portrait of the PI3K/AKT pathway in colorectal cancer. Biochim Biophys Acta 2015; 1855: 104-121.
- Khamas A, Ishikawa T, Shimokawa K, Mogushi K, Iida S, Ishiguro M, Mizushima H, Tanaka H, Uetake H, Sugihara K. Screening for epigenetically masked genes in colorectal cancer Using 5-Aza-2'-deoxycytidine, microarray and gene expression profile. Cancer Genomics Proteomics 2012; 9: 67-75.
- Ruivo CF, Adem B, Silva M, Melo SA. The biology of cancer exosomes: insights and new perspectives. Cancer Res 2017; 77: 6480-6488.
- Kalluri R. The biology and function of exosomes in cancer. J Clin Invest 2016; 126: 1208-1215.
- Issa IA, Noureddine M. Colorectal cancer screening: An updated review of the available options. World J Gastroenterol 2017; 23: 5086-5096.
- Zhu M, Huang Z, Zhu D, Zhou X, Shan X, Qi LW, Wu L, Cheng W, Zhu J, Zhang L, Zhang H, Chen Y, Zhu W, Wang T, Liu P. A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget 2017; 8: 17081-17091.
- Peach CJ, Mignone VW, Arruda MA, Alcobia DC, Hill SJ, Kilpatrick LE, Woolard J. Molecular pharmacology of VEGF-A isoforms: binding and signalling at VEGFR2. Int J Mol Sci 2018; 19: 1264.