ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 7
Analysis of R1587K polymorphism of ATP-binding cassette transporter A1 gene among type 2 diabetes mellitus Malaysian subjects
1Genetic Research Group, Department of Biomedical Science, Faculty of Medicine and Health Sciences, Universiti Putra Malaysia, Serdang, Selangor, Malaysia
2Malaysian Research Institute of Aging, Universiti Putra Malaysia, Serdang, Selangor, Malaysia
#These authors contributed equally to this work
- *Corresponding Author:
- Polin Haghvirdizadeh
Department of Biomedical Science
Genetic Research Group
Faculty of Medicine and Health Sciences
Universiti Putra Malaysia, Malaysia
Accepted on December 7, 2016
Objective: Impaired insulin resistance, insulin secretion and dysregulation of lipid and protein metabolism followed by environmental and genetic factors cause to Type 2 Diabetes Mellitus (T2DM) which is a complex polygenic disorder. Several studies have been reported that ATP Binding Cassette Transporter A1 (ABCA1) gene polymorphisms in known as one of the genetic risk factor for T2DM. This study was conducted to determine the frequency of ABCA1 gene by rs2230808 (R1587K) polymorphism in Malaysian subjects.
Methods: The genotype for ABCA1 gene polymorphism was determined based on high resolution melting-PCR among 164 T2DM and 165 control subjects.
Result: There was a significant difference between the subjects in terms of FPG (p value=0.036*). Regarding genotypic and allelic frequency, we could not find any significance in this polymorphism among subjects. The frequency of Wild Type (WT) genotypes among T2DM and control subjects was 56.7% and 61.8% respectively. In addition, the WT and Homozygous (HOM) genotypes were significantly higher in controls (p value=0.008 and p value=0.006 respectively) and the Heterozygous (HET) genotypes were higher in T2DM subjects (p value=0.001).
Conclusion: The R1587K polymorphism of ABCA1 gene cannot be considered as a genetic risk factor for T2DM among Malaysian subjects.
Keywords
ABCA1, Polymorphism, Type 2 diabetes mellitus.
List of Abbreviations
ABCA1: ATP-Binding Cassette Transporter A1; ADA: American Diabetes Association; BMI: Body Mass Index CI: Confidence Interval; DBP: Diastolic Blood Pressure; FPG: Fasting Plasma Glucose; HDL: High Density Lipoprotein; HET: Heterozygous; HOM: Homozygous IFG/IGT: Impaired Fasting Glucose/Impaired Glucose Tolerance; LDL: Low Density Lipoprotein; NCDs: Non-Communicable Diseases; PCR-HRM: Polymerase Chain Reaction-High Resolution Melting; SBP: Systolic Blood Pressure; T2DM: Type 2 Diabetes Mellitus; TG: Triglyceride; WHO: World Health Organization; WHR: Waist Hip Ratio; WT: Wild Type.
Introduction
Prediabetes known as a middle state of hyperglycaemia with glycaemic parameters. Today different organizations have been describing the prediabetes with different criteria’s that is not shown as a uniform pattern [1]. According to the American Diabetes Association (ADA), the terms of pre-diabetes refers to the metabolic clinical condition that is able to influence an individual for the future development of diabetes [2]. Also, World Health Organization (WHO) definition for prediabetes is a state of intermediate hyperglycaemia using two specific parameters, impaired Fasting Glucose (IFG) and/or Impaired Glucose Tolerance (IGT) or a combination of the two based on a 2 hours Oral Glucose Tolerance Test (OGTT) [1]. Such evidences suggest association between prediabetes and complications of diabetes such early nephropathy, small fiber neuropathy, early retinopathy and risk of macrovascular disease [1].
One of the four preferences Non-Communicable Diseases (NCDs), which identified by the WHO is diabetes [3]. In Malaysia, based on the latest National Health and Morbidity Survey in 2015, the prevalence of diabetes mellitus was reported as 17.5% [4]. Diabetes has become the global leading causes of mortality and morbidity due to its complications [5].
The prevalence of T2DM is increasing to 300 million people by the year 2025 [6]. Impaired insulin resistance, insulin secretion and dysregulation of lipid and protein metabolism with environmental and genetic factors cause to T2DM which is a complex polygenic disorder [7]. The role of genetic polymorphisms involved in diabetes is still remaining unclear; however, several association researches suggested various candidate genes for T2DM in different populations [8]. To provide the supporting evidence for the role of gene polymorphism in individuals, can refer the subjects to the genetic and allelic frequency of polymorphism among studied groups. The genetic polymorphisms of ABCA1 gene provide a basis for studying the association between genetic variants and the development of T2DM in different ethnic groups. Transport of cholesterol and phospholipids from cells to lipid-poor apolipoproteins mediated by ABCA1 gene, which located on the chromosome 9 region q31.1 [9]. This study was conducted to determine genetic polymorphism of the rs2230808 (R1587K) among T2DM subjects.
Material and Methods
Study subjects
Ethical approval has been obtained from the ethical committee of the National heart Institute, Malaysia (Ref No. IJNEC/05/10 (02)). A total of 164 T2DM subjects were collected from National Heart Institute, Kuala Lumpur. Subjects were selected as (adults>30 years old) who has been diagnosed with T2DM. A questionnaire in both Malay and English languages were obtained to assess the socio-demographic factors. Informed consent was obtained from all the subjects who have participated in this study. A total of 165 control individuals were recruited from the healthy respondents who didn’t haveT2DM at the time of sample collection. All the subjects has been selected based on the inclusion/exclusion criteria and those who had been diagnosed with any type of cancer, type 1 diabetes, genetic malformation and pregnancy were excluded from this study.
Genomic DNA extraction
The buccal and blood specimens were collected from the case and control subjects respectively. The genomic DNA was extracted by using the commercial buccal and blood extraction kits (QIGEN) and was stored at-20ᴼC for further analysis. PCR optimization was carried out for all primers, followed by DNA qualifications (gel electrophoresis) and qualification (Nano drop in two OD wave lengths 260 nm and 280 nm) before amplification of the candidate genes.
Genotyping of polymorphisms
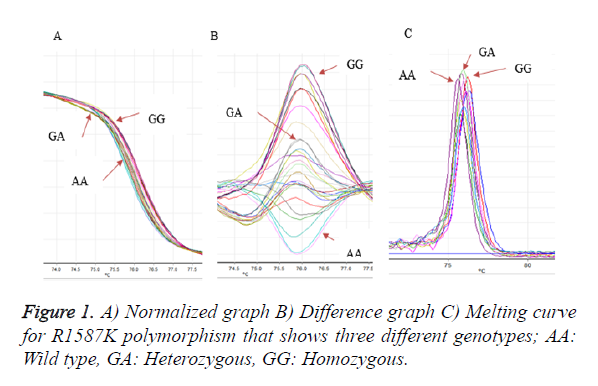
Genotyping of this polymorphism was done based on real time PCR-HRM and the primers were designed and synthesized by NextGene Sdn Bhd. The PCR products of samples that were amplified from the respective genes were sequenced and confirmed with the databases. In each cycling condition, the positive control for the respective genes followed by the negative control consisted of PCR-grade water lacking the DNA template was used accordingly. All samples were genotyped by high resolution melting analysis by Rotor Gene 6000. PCR primers for R1587K were forward primer CTACTAGAACAAAGACAGCGGTTTACC and reverse primer CGATTTCTCAACAGCTTGGGAAGAT. PCR was performed in a volume of 25 μL consisting of about 1 μL of genomic DNA, 0.5 μL each of forward and reverse primers, 13 μL water and 10 μL of master mix for HRM (Epitect HRM Mastermix, QIGEN). The temperature was kept hold at 95 for 5 min for denaturation followed by 40 cycles of 20 s at 95°C, 10 s at 55°C, and 20 s at 72°C. The melting curves were normalized (temperature ranges on each side of the melting transition were chosen and the data points for a given sample were scaled between 0 and 100% fluorescent intensity). Based on used control samples (wild type (WT) and Homozygous mutant (HOM)) were confirmed by sequencing. Table 1 demonstrated the sequence and length and Tm of each primer.
| T2DM | Control | P value | |||||
|---|---|---|---|---|---|---|---|
| WT (AA) | HET (GA) | HOM (GG) | WT (AA) | HET (GA) | HOM (GG) | ||
| Age (Years) | 61.83 ± 8.98 | 62.67 ± 10.59 | 61.50 ± 8.12 | 54.69 ± 12.07 | 57.29 ± 10.22 | 49.93 ± 13.23 | 0.24 |
| BMI (Kg/m2) | 27.90 ± 4.94 | 27.97 ± 5.43 | 26.61 ± 5.52 | 27.16 ± 7.25 | 27.02 ± 4.19 | 26.66 ± 4.51 | 0.767 |
| WHR | 0.97 ± 0.20 | 0.95 ± 0.06 | 0.90 ± 0.15 | 0.97 ± 0.70 | 0.92 ± 0.09 | 0.91 ± 0.07 | 0.791 |
| SBP (mmHg) | 145.26 ± 23.71 | 137.21 ± 25.54 | 139.57 ± 18.12 | 137.48 ± 19.41 | 138.89 ± 20.63 | 137.56 ± 24.96 | 0.479 |
| DBP (mmHg) | 79.87 ± 12.74 | 76.26 ± 11.60 | 76.57 ± 5.56 | 79.17 ± 10.01 | 79.95 ± 9.87 | 80.75 ± 15.23 | 0.481 |
| FPG (mmol/L) | 8.58 ± 3.53 | 8.99 ± 3.54 | 7.06 ± 2.77 | 5.50 ± 1.30 | 5.52 ± 1.39 | 4.91 ± 1.01 | 0.036* |
| HbA1c (mmol/L) | 7.97 ± 2.14 | 8.02 ± 2.27 | 7.75 ± 2.25 | 5.82 ± 0.61 | 6.02 ± 0.78 | 6.04 ± 0.50 | 0.438 |
| LDL (mmol/L) | 2.32 ± 0.97 | 2.32 ± 0.74 | 2.92 ± 1.10 | 2.67 ± 0.91 | 2.47 ± 0.94 | 2.73 ± 1.22 | 0.138 |
| HDL (mmol/L) | 1.28 ± 0.42 | 1.24 ± 0.57 | 1.17 ± 0.29 | 1.24 ± 0.48 | 1.14 ± 0.35 | 1.15 ± 0.23 | 0.382 |
| TG (mmol/L) | 1.84 ± 1.47 | 1.70 ± 0.74 | 1.33 ± 0.31 | 1.25 ± 0.68 | 1.42 ± 0.93 | 1.01 ± 0.67 | 0.139 |
| Chol (mmol/L) | 4.41 ± 1.27 | 4.29 ± 0.93 | 4.70 ± 1.19 | 4.51 ± 1.06 | 4.22 ± 1.24 | 4.58 ± 1.53 | 0.218 |
| One Way ANOVA test, Values shown are the mean ± SD, *P<0.05 | |||||||
Table 1. Clinical and biochemical characteristics according to the genotypes of the subjects.
Statistical analysis
To analyse the data in this study, the Statistical Package for Social Science (SPSS) (Version 21) was assisted. Descriptive statistics were utilized to analyse all variable information such as socio-demographic and anthropometric factors followed by the genotypes of all the study subjects. Moreover, all of these factors were compared by using Student’s t-test, and one way ANOVA test to compare the group means and a level of p<0.05 was considered as a statistically significant. Confidence intervals (95%) were reported where appropriate. Allelic frequencies were calculated by the gene counting method and the genotype distribution with Hardy-Weinberg by assisting the chi-squared test.
Results
R1587K
Analysis of R1587K polymorphism with 86 bp PCR product with mutation (A → G) was amplified by PCR-HRM. Three different genotypes of R1587K were detected through this method with special difference between the shapes of each curve. After running a few samples with different shifting curves confirm those genotyping patterns by sequencing and use them as a reference samples. Figure 1 showed three different graphs that has been detected during the genotyping of R1587K polymorphism of ABCA1gene (homozygous, heterozygous and wildtype).
In this study 329 subjects were participated that were divided into 2 groups: 165 control and 164 T2DM subjects. The association of anthropometric and social demographic factors and major risk factors in T2DM with ABCA1 polymorphism among Malaysian subjects was investigated.
Table 1 demonstrated the mean of clinical and biochemical characteristics among T2DM and control subjects based on 3 different genotypes for R1587K polymorphism. As shown in this table the mean of age for HOM GG genotype represented the youngest groups (61.50 ± 8.12 and 49.93 ± 13.23) for T2DM and control respectively. The same pattern was observed for the BMI in the HOM GG pattern that was the lowest values in both groups (26.61 ± 5.52 and 26.66 ± 4.51) respectively followed by the WHR as the lowest value for HOM GG genotype (0.90 ± 0.06 and 0.91 ± 0.07) respectively. For the SBP, the highest values were presented in WT AA genotype in diabetes group (145.26 ± 23.71) but interestingly represented the lowest value among the healthy individuals (137.48 ± 19.41) respectively. The FPG values was following the same pattern as BMI, WHR and age which represented the lowest values among HOM GG genotype for both T2DM and control groups (7.06 ± 2.77 and 4.91 ± 1.01) respectively and represented as the only significant value among the biochemical factors (P ≤ 0.05). Moreover, the values for the LDL (2.92 ± 1.10 and 2.73 ± 1.22) were not following the BMI and WHR values and showed the highest values among the three genotypes of T2DM and control individuals. The HDL values was following the SBP and represented as the highest values in both T2DM and healthy group in WT AA genotype as the highest values (1.28 ± 0.42 and 1.24 ± 0.48) respectively. The TG values in both diabetes and healthy groups were (1.33 ± 0.31 and 1.01 ± 0.67) respectively in HOM GG genotypes as the lowest values that were following the BMI and WHR. The last but not the least, the values for the Cholesterol was represented in the HET GA genotype as the lowest values in both diabetes and healthy groups (4.29 ± 0.93 and 4.22 ± 1.24) respectively.
The overall significant difference were not observed among the genotype and allelic frequency of T2DM and control subjects (NS) and (p value=0.965) respectively (Table 2). The genotypic and allelic frequencies of R1587K polymorphism among subjects, the percentage of WT, HET and HOM among T2DM subjects was 56.7%, 38.4% and 4.8% respectively and the percentage of WT, HET and HOM in control subjects was 61.8%, 28.4% and 9.6% respectively. In addition, WT, HET and HOM showed the significant difference between T2DM and control subjects which showed significantly WT and HOM were higher in controls and HET was higher in T2DM (p value=0.008, p value=0.006 and p value=0.001 respectively).
| Genotypes and alleles | T2DM (%) | Controls (%) | P-value | |
|---|---|---|---|---|
| WT (AA) | 93 (56.7%) | 102 (61.8%) | ||
| HET (GA) | 63 (38.4%) | 47 (28.4%) | NS | |
| HOM (GG) | 8 (4.8%) | 16 (9.6%) | ||
| Alleles | ||||
| A | 249 (75.9%) | 251 (76%) | ||
| G | 79 (24.1%) | 79 (24%) | 0.965 | |
| Post hoc test | (95% CI) | P-value | ||
| AA vs. GA | -0.23 | 0.108 | ||
| AA vs. GG | -0.41 | 0.184 | ||
| GA vs. GG | 0.01-0.46 | 0.034* | ||
| *P value<0.05, between T2DM and controls, WT: Wild Type; HOM: Homozygous; HET: Heterozygous. | ||||
Table 2. Genotypic and allelic frequencies of R1587K polymorphism among subjects.
Also, the percentage of A allele was 75.9% and 76% respectively among the T2DM and controls and the percentage of G allele was 24.1% and 24% respectively. In addition, post hoc test demonstrated the significant difference between HET genotype and HOM GG genotype among the subjects (p value=0.034) but no difference between other genotypes.
Discussion
However, in other populations, did not work on the association of this polymorphism with T2DM; in current study, there was no significant difference between genotypes and alleles of R1587K polymorphism among T2DM and control subjects [10].
Conclusion
The present study could not show a significant genetic association for R1587K polymorphism of ABCA1 gene among Malaysian Malay T2DM patients compared to control subjects. Replication studies with larger number of samples on a homogenous study population are strongly recommended to confirm the association of ABCA1 gene polymorphisms with T2DM.
Competing Interest
The author(s) declare that they have no competing interests.
Authors' Contributions
Polin Haghvirdizadeh conceived the study and participated in the experimental design, data acquisition and analysis, interpretation of results, and drafted the manuscript. Polin Haghvirdizadeh, Ali Etemad, Vasudevan Ramachandran and Patimah Ismail interpreted the results and critically reviewed the study for important intellectual content. All authors approved the final version of the manuscript.
Acknowledgements
The authors would like to extend their gratitude to all the volunteers who involved in this study.
References
- Bansal N. Prediabetes diagnosis and treatment: A review. W J Diab 2015; 6: 296.
- Ciccone MM, Scicchitano P, Cameli M, Cecere A, Cortese F, Dentamaro I, Gentile F, Gesualdo M, Maiello M, Modesti PA, Muiesan ML. Endothelial function in pre-diabetes, diabetes and diabetic cardiomyopathy: a review. J Diab Metabol 2014; 2014.
- Unwin N, Gan D, Whiting D. The IDF Diabetes Atlas: providing evidence, raising awareness and promoting action. Diab Res Clin Pract 2010; 87: 2-3.
- Hilmi A. Alarming increase in diabetes among Malaysians-Nation-The Star Online. Thestar.com.my 2016.
- Whiting DR, Guariguata L, Weil C, Shaw J. IDF diabetes atlas: global estimates of the prevalence of diabetes for 2011 and 2030. Diab Res Clin Pract 2011; 94: 311-321.
- Zaharna MM, Abed AA, Sharif FA. Calpain-10 gene polymorphism in type 2 diabetes mellitus patients in the Gaza Strip. Med Princip Pract 2010; 19: 457-462.
- Moon SS, Lee JE, Lee YS, Kim SW, Jeoung NH, Lee IK, Kim JG. Association of pyruvate dehydrogenase kinase 4 gene polymorphisms with type 2 diabetes and metabolic syndrome. Diab Res Clin Pract 2012; 95: 230-236.
- Plengvidhya N. Molecular genetics of diabetes mellitus. Siriraj Med J 2008; 60: 273-278.
- Kolovou V, Kolovou G, Marvaki A, Karakosta A, Vasilopoulos G, Kalogiani A, Degiannis D, Marvaki C, Demopoulos CA. ATP-binding cassette transporter A1 gene polymorphisms and serum lipid levels in young Greek nurses. Lip Health Dis 2011; 10: 1.
- Villarreal-Molina T, Posadas-Romero C, Romero-Hidalgo S, Antunez-Arguelles E, Bautista-Grande A, Vargas-Alarcon G, Kimura-Hayama E, Canizales-Quinteros S, Juárez-Rojas JG, Posadas-Sanchez R, Cardoso-Saldana G. The ABCA1 gene R230C variant is associated with decreased risk of premature coronary artery disease: the genetics of atherosclerotic disease (GEA) study. PLoS One 2012; 7: e49285.