ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 16
The single nucleotide polymorphism of DMRT1 is associated with oligospermia
1Department of Reproductive Medicine, the First Affiliated Hospital of Dali University, Dali, People’s Republic of China
2Clinical Medicine College of Dali University, Dali, People’s Republic of China
3Department of Reproductive and Genetic, the First Affiliated Hospital of Kunming Medical University, Kunming, People’s Republic of China
4Department of Biochemistry and Molecular Biology, College of Basic Medical Sciences, Dali University, Dali, People’s Republic of China
#These authors contributed equally to this work.
- *Corresponding Author:
- Shu-Hua Zhao
Department of Reproductive and Genetic
The First Affiliated Hospital of Kunming Medical University
Kunming, People’s Republic of China
- *Corresponding Author:
- Wei Xiong
Department of Biochemistry and Molecular Biology
College of Basic Medical Sciences, Dali University
Dali, People’s Republic of China
Accepted date: July 24, 2017
Male infertility is a common human reproductive defect that can be resulted from spermatogenesis impairment. DMRT1 encodes a male-specific transcriptional regulator and is important for human spermatogenesis. This study was designed to explore the relationship between the Single Nucleotide Polymorphism (SNP) of DMRT1 and oligospermia. Two SNPs namely rs3739583 and rs4742608, were screened in 212 infertile men with oligospermia and 217 normal controls by using a PCR-RFLP assay. The genotyping data showed that the frequencies of allele A (64.9% vs. 57.8%, p=0.041, OR=1.346, 95% CI 1.021~1.773) and genotype AA (39.1% vs. 29.0%, p=0.027, OR=1.604, 95% CI 1.073~2.398) of SNP rs4742608 were significantly higher in the infertile patients with oligospermia compared with the normal controls. Our data suggest that the SNP rs4742608 in DMRT1 is associated with oligospermia and may affect the genetic susceptibility to human oligospermia.
Keywords
DMRT1, Single nucleotide polymorphism (SNP), Male infertility, Spermatogenesis, Oligospermia.
Introduction
Male infertility is a common human reproductive defect that accounts for about half of infertile cases [1,2]. A number of genetic factors are involved in male infertility, among which spermatogenesis impairment is one of the most common causes [3-5]. Spermatogenesis is a complex and highly regulated process involving the expression of thousands of genes and the alterations of these genes may cause spermatogenesis impairment [6-14]. Thus, the genes involved in spermatogenesis may be important candidates for spermatogenesis impairment and male infertility. Over the last decades, some genetic abnormalities leading to spermatogenesis impairment, such as chromosomal aberrations, deletions in the AZF regions of Y chromosome and mutations of certain genes, have been identified [15,16]. However, these known genetic abnormalities can only explain 25-30% cases of spermatogenesis impairment [17]. More genes related to spermatogenesis impairment remain to be identified. Human doublesex and mab-3 related transcription factor 1 (DMRT1) is localized on chromosome 9p24.3 and is homologous to the male sexual regulatory gene mab-3 from the nematode Caenorhabditis elegans and the Drosophila melanogaster sex regulatory gene double sex [18]. This gene encodes a male-specific transcriptional regulator containing a conserved zinc finger-like DNA-binding domain (DM domain) and a nuclear localization signal, and plays a key role in sex determination and differentiation. DMRT1 is mainly expressed in testis including sertoli cells, spermatogonia and spermatocytes, suggesting that DMRT1 may play an important role in human spermatogenesis [18,19]. In recent year, accumulating evidences demonstrate that human DMRT1 may be important for gametogenesis and male reproduction [20]. In mice, DMRT1 is required for the progression of mitosis and meiosis of male germ cells and deletion of DMRT1 results in spermatogenesis failure and male infertility, indicating that DMRT1 is critical for spermatogenesis of mice [21-24].
Currently, little is known about whether DMRT1 is involved in human spermatogenesis impairment. In this study, we selected two common exonic SNPs (rs3739583 and rs4742608) of DMRT1 from dbSNP and investigated the relationship between these two SNPs and human spermatogenesis in 212 infertile patients with oligospermia and 217 controls in Chinese population.
Materials and Methods
212 infertile patients with idiopathic oligospermia (sperm number less than 15 × 106/ml), aged from 24-41 y old, were enrolled in this study. All patients underwent semen analyses at least twice according to the WHO guidelines [25]. Patients with diseases known to affect spermatogenesis, such as orchitis, maldescensus of testis, varicocele and obstruction of vas deferens, were excluded. In addition, patients with chromosomal abnormalities and microdeletions of AZF region on Y chromosome were also excluded after genetic analysis [26]. 217 fertile men aged from 24-48 y old with normal semen profile, were served as normal controls. These men had at least one offspring without technical assistance of reproduction.
This study was approved by the Institutional Review Board of Affiliated Hospital of Dali University and informed approval was obtained from all participants.
PCR amplification
DNA was extracted from the peripheral blood leucocytes of patients and controls using a TIAN amp Genomic DNA Kit (TIANGEN, Beijing, China). Two pairs of primers were designed to amplify the DNA fragments including the SNPs rs3739583 and rs4742608. Because there was no restriction enzyme site for SNP rs4742608, the forward primer with a mismatched nucleotide was used to generate a restriction site AccI. The sequences of primers and the expected size of PCR products are shown in Table 1. PCR was performed in a total volume of 25 μl containing 100 ng of genomic DNA, 200 μmol/L dNTPs, 1.5 mmol/L MgCl2, 10 pmol of each primer, 2.5 μl of 10 × PCR buffer and 1 U Taq polymerase (Takara, Shiga, Japan). The PCR running condition was: 94°C for 5 min, 94°C for 30 s, 58°C for 30 s, 72°C for 40 s, 35 cycles, and 72°C for 5 min.
| Primer sequence* | Size | Enzyme | Sizes of alleles |
|---|---|---|---|
| rs3739583 | 325 bp | TaqI | Allele T: 202 bp+123 bp |
| F: 5'-TTCATCCCTCGCAGCAGTCT-3' | Allele A: 325 bp | ||
| R: 5'-AGGCGTAGCCGTGGTTCCT-3’ | |||
| rs4742608 | AccI | Allele A: 233 bp+27 bp | |
| F: 5'-GAAACTAGTCTAAAAAAATTCATTGGTCT-3' | 260 bp | Allele G: 260 bp | |
| R: 5'-CTAAAACCACTGGTGGATGA-3' |
Table 1: Primers, size of PCR product, restriction enzymes and size of alleles for RFLP analysis.
Genotyping
A Restriction Fragment Length Polymorphism (RFLP) assay was performed to genotype SNP rs3739583 and rs4742608. PCR products were digested overnight with corresponding restriction enzymes (Fermentas, Vilnius, Lithuania) according to the manufacturer’s protocols and then electrophoresed on a 3% agarose gel. Restriction enzymes and the size of digested fragments of alleles were shown in Table 1. The genotypes were further confirmed by direct sequencing of the PCR products.
Statistical analysis
The allele and genotype frequencies of rs3739583 and rs4742608 in patients and controls were manually quantified. The Hardy-Weinberg equilibrium was tested by using Hardy- Weinberg equilibrium calculator [27]. The differences in allelic and genotypic frequencies of the two SNPs between patients and controls were assessed by chi-square test. Odds Ratios (ORs), 95% Confidence Intervals (CIs) and specificity were calculated by using unconditional logistic regression analyses. The difference between two groups was considered to be statistically significant if the p value was smaller than 0.05. All statistical analyses were performed by using IBM SPSS 19.
Results
The polymorphic distributions of SNPs rs3739583 and rs4742608 in DMRT1 were analyzed for 212 infertile patients with oligospermia and 217 fertile controls by using a PCRRFLP assay. The frequency distributions of the allele and genotype of the two SNPs were shown in Table 2. The genotypic distributions of the two SNPs were consistent with the Hardy-Weinberg equilibrium in both patients and normal controls (data not shown). As shown in Table 2, there was no significant difference in frequencies of allele and genotype of SNP rs3739583 between patients and controls.
| SNP | Genotype/Allele | Controls (n=217) | Patients (n=212) total | P value (OR, CI% 95)a |
|---|---|---|---|---|
| Azoospermia | ||||
| Oligozoospermia | ||||
| rs3739583 | TT | 0.273 (59) | 0.264 (56) | 0.943 (0.961, 0.627~1.474) |
| TA | 0.483 (105) | 0.533 (113) | 0.357 (0.833~1.779,1.218) | |
| AA | 0.244 (53) | 0.203 (43) | 0.361 (0.787. 0.499~1.242) | |
| T | 0.514 (223) | 0.531 (225) | 0.671 (0.935, 0.715~1.222) | |
| A | 0.486 (211) | 0.469 (199) | ||
| rs4742608 | AA | 0.290 (63) | 0.391 (84) | 0.027 (1.604, 1.073~2.398) |
| AG | 0.576 (125) | 0.505 (107) | 0.166 (0.75, 0.513~1.100) | |
| GG | 0.134 (29) | 0.104 (21) | 0.334 (0.713, 0.393~1.294) | |
| A | 0.578 (251) | 0.649 (275) | 0.041 (1.340, 1.021~1.773) | |
| G | 0.422 (183) | 0.351 (149) |
Table 2: The allele and genotype frequency of the SNPs in patients and controls.
The significant difference in the polymorphic distribution of rs4742608 was detected between the patients with oligospermia and controls. The frequencies of allele A (64.9% vs. 57.8%, p=0.044, OR=1.346, 95% CI 1.021~1.773) and genotype AA (39.1% vs. 29.0%, p=0.027, OR=1.604, 95% CI 1.073~2.398) at rs4742608 locus were significantly higher in the patients with oligospermia than those in the controls.
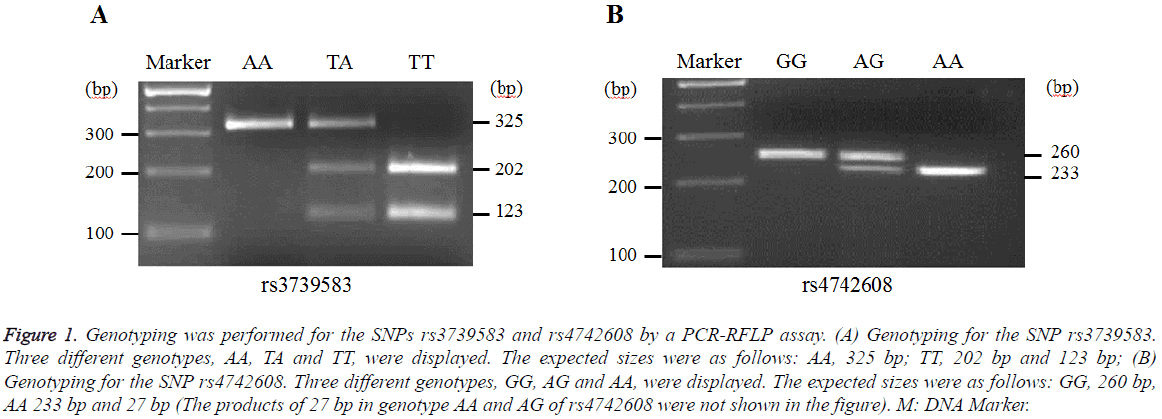
Representative genotyping images for the SNPs rs3739583 and rs4742608 in DMRT1 were shown in Figure 1. The genotypes of AA, TA, TT and GG, AG, AA were displayed for rs3739583 and rs4742608, respectively.
Figure 1: Genotyping was performed for the SNPs rs3739583 and rs4742608 by a PCR-RFLP assay. (A) Genotyping for the SNP rs3739583. Three different genotypes, AA, TA and TT, were displayed. The expected sizes were as follows: AA, 325 bp; TT, 202 bp and 123 bp; (B) Genotyping for the SNP rs4742608. Three different genotypes, GG, AG and AA, were displayed. The expected sizes were as follows: GG, 260 bp, AA 233 bp and 27 bp (The products of 27 bp in genotype AA and AG of rs4742608 were not shown in the figure). M: DNA Marker.
Discussion
The genetic causes of spermatogenesis impairment are very complex and the majority of genes involved in spermatogenesis impairment remains unknown. By using the gene knockout technique, hundreds of mouse models with spermatogenesis impairment have been established and a number of candidate genes for human spermatogenesis impairment were identified [28,29]. Given the genetic background difference between mouse and human, whether these candidate genes also regulate spermatogenesis in human needs to be further investigated. The polymorphisms or variations in candidate genes are potential risk factors for human spermatogenesis impairment. Thus, the polymorphic analysis in candidate genes represents an interesting area of research to investigate the relationship between the candidate gene and spermatogenesis impairment in human [17,30].
DMRT1 is an important candidate gene for spermatogenesis impairment and its role in spermatogenesis impairment in mice has been proven [24]. In recent years, the role of this gene in human spermatogenesis impairment has caught more attention. A recent study showed that the coding sequence deletion of DMRT1 was detected in some men with azoospermia, suggesting that the genetic abnormality of DMRT1 is associated with azoospermia and affects spermatogenesis in human [31]. Thus, it was speculated that the polymorphism of DMRT1 might be also involved in human oligospermia. To test this hypothesis, we investigated the polymorphic distributions of the SNPs rs3739583 and rs4742608 in DMRT1 in patients with oligospermia and normal controls in this study. As the result shown, the polymorphic distribution of SNP rs3739583 had no significant difference between patients with oligospermia and normal controls, suggesting that the polymorphism of SNP rs3739583 is not associated with oligospermia and this SNP may not affect the susceptibility to oligospermian. However, the allelic frequency and genotype of rs4742608 had significant difference between the patients with oligospermian and the controls. The frequencies of allele A and genotype AA were significantly higher in patients than those in controls, indicating that the polymorphism of SNP rs4742608 is associated with oligospermia, and genotype AA may increase the risk of oligospermia (OR=1.6). These results suggest that SNP rs4742608 may affect the susceptibility to oligospermia. The SNP rs4742608 is located within the 3′- untranslated region (3’ UTR) of DMRT1. Thus, it is possible that this SNP affects DMRT1 protein translation and then causes abnormal spermatogenesis. Of course, whether this SNP does really affect the translation of DMRT1 needs to be further investigated. It cannot be excluded that the SNP rs4742608 is only a genetic marker of oligospermia, which is in linkage disequilibrium with other locus that plays a role in oligospermia.
Conclusion
In this study, we first report the relationship between the polymorphism of DMRT1 and human oligospermia. Our results indicated that polymorphism of SNP rs4742608 in DMRT1 is associated with oligospermia and may affect the susceptibility to oligospermia in Chinese population, suggesting that DMRT1 may be involved in human spermatogenesis impairment. However, our findings need to be validated in further studies with larger size of sample and other ethnic populations, since the sample size in this study is limited and also restricted to Chinese population.
Acknowledgment
This work was supported by Doctoral Degree Activation Fund for Scientific Research Projects of Dali University (KYBS201612), Research topic for Dali Bai Autonomous Prefecture Committee of Jiusan Society (16DJ04) and National Natural Science Foundation of China (81300542, 81560458 and 31601155).
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- De Kretser DM, Baker HW. Infertility in men: recent advances and continuing controversies. J Clin Endocrinol Metabol 1999; 84: 3443-3450.
- Zhang Y, Xiao F, Lu S, Song J, Zhang C, Li J, Gu K, Lan A, Lv B, Zhang R, Mo F, Jiang G, Zhang X, Yang X. Research trends and perspectives of male infertility: a bibliometric analysis of 20 years of scientific literature. Androl 2016.
- Toshimori K, Ito C, Maekawa M, Toyama Y, Suzuki-Toyota F, Saxena DK. Impairment of spermatogenesis leading to infertility. Anatom Sci Int 2004; 79: 101-111.
- Massart A, Lissens W, Tournaye H, Stouffs K. Genetic causes of spermatogenic failure. Asian J Androl 2012; 14: 40-48.
- Neto FT, Bach PV, Najari BB, Li PS, Goldstein M. Genetics of male infertility. Curr Urol Rep 2016; 17: 70.
- Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Nat Acad Sci US Am 2003; 100: 12201-12206.
- Neto FT, Bach PV, Najari BB, Li PS, Goldstein M. Spermatogenesis in humans and its affecting factors. Semin Cell Dev Biol 2016.
- Stouffs K, Tournaye H, Liebaers I, Lissens W. Male infertility and the involvement of the X chromosome. Hum Reprod Update 2009; 15: 623-637.
- Inoue S, Tomasini R, Rufini A, Elia AJ, Agostini M, Amelio I, Cescon D, Dinsdale D, Zhou L, Harris IS, Lac S, Silvester J, Li WY, Sasaki M, Haight J, Brustle A. TAp73 is required for spermatogenesis and the maintenance of male fertility. Proc Nat Acad Sci US Am 2014; 111: 1843-1848.
- Lu C, Xu M, Wang Y, Qin Y, Du G, Wu W, Han X, Ji C, Yang Y, Gu A, Xia Y, Song L, Wang S, Wang X. Genetic variants in meiotic program initiation pathway genes are associated with spermatogenic impairment in a Han Chinese population. PloS One 2013; 8: e53443.
- Xu XB, Liu SR, Ying HQ, A ZC. Null genotype of GSTM1 and GSTT1 may contribute to susceptibility to male infertility with impaired spermatogenesis in Chinese population. Biomarkers 2013; 18: 151-154.
- Ying HQ, Scott MB, Zhou-Cun A. Relationship of SNP of H2BFWT gene to male infertility in a Chinese population with idiopathic spermatogenesis impairment. Biomarkers 2012; 17: 402-406.
- Cheng P, Chen H, Liu SR, Pu XY, A ZC. SNPs in KIT and KITLG genes may be associated with oligospermia in Chinese population. Biomarkers 2013; 18: 650-654.
- Ying HQ, Pu XY, Liu SR, A ZC. Genetic variants of eNOS gene may modify the susceptibility to idiopathic male infertility. Biomarkers 2013; 18: 412-417.
- O'Flynn O'Brien KL, Varghese AC, Agarwal A. The genetic causes of male factor infertility: a review. Fertility Sterility 2010; 93: 1-12.
- Dhanoa JK, Mukhopadhyay CS, Arora JS. Y-chromosomal genes affecting male fertility: A review. Veterinary World 2016; 9: 783-791.
- Ferlin A, Raicu F, Gatta V, Zuccarello D, Palka G, Foresta C. Male infertility: role of genetic background. Reprod Biomed Online 2007; 14: 734-745.
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D. Evidence for evolutionary conservation of sex-determining genes. Nature 1998; 391: 691-695.
- Ying M, Chen B, Tian Y, Hou Y, Li Q, Shang X, Sun J, Cheng H, Zhou R. Nuclear import of human sexual regulator DMRT1 is mediated by importin-beta. Biochimica et biophysica acta 2007; 1773: 804-813.
- Zarkower D. DMRT genes in vertebrate gametogenesis. Curr Topics Dev Biol 2013; 102: 327-356.
- Kim S, Namekawa SH, Niswander LM, Ward JO, Lee JT, Bardwell VJ, Zarkower D. A mammal-specific doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS Genet 2007; 3: e62.
- Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, Zarkower D. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell 2010; 19: 612-624.
- Raymond CS, Murphy MW, O'Sullivan MG, Bardwell VJ, Zarkower D. DMRT1, a gene related to worm and fly sexual regulators is required for mammalian testis differentiation. Genes Dev 2000; 14: 2587-2595.
- Agbor VA, Tao S, Lei N, Heckert LL. A Wt1-DMRT1 transgene restores DMRT1 to sertoli cells of DMRT1(-/-) testes: a novel model of DMRT1-deficient germ cells. Biol Reprod 2013; 88: 51.
- World Health Organization. Laboratory manual of the WHO for the examination of human semen and sperm-cervical mucus interaction. Ann Ist Super Sanita 2001; 37: 1-123.
- Simoni M, Bakker E, Krausz C. EAA/EMQN best practice guidelines for molecular diagnosis of y-chromosomal microdeletions: State of the art 2004. Int J Androl 2004; 27: 240-249.
- Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol 2009; 169: 505-514.
- O'Bryan MK, de Kretser D. Mouse models for genes involved in impaired spermatogenesis. Int J Androl 2006; 29: 76-89.
- Maduro MR, Lamb DJ. Understanding new genetics of male infertility. J Urol 2002; 168: 2197-2205.
- Nuti F, Krausz C. Gene polymorphisms/mutations relevant to abnormal spermatogenesis. Reprod Biomed Online 2008; 16: 504-513.
- Lopes AM, Aston KI, Thompson E, Carvalho F, Goncalves J, Huang N, Matthiesen R, Noordam MJ, Quintela I, Ramu A, Seabra C, Wilfert AB, Dai J, Downie JM, Fernandes S, Guo X. Human spermatogenic failure purges deleterious mutation load from the autosomes and both sex chromosomes, including the gene DMRT1. PLoS Genet 2013; 9: e1003349.