ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Special Issue Article - Biomedical Research (2018) Medical Diagnosis and Study of Biomedical Imaging Systems and Applications
The influence of miR-366 on colorectal cancer cell proliferation and migration by targeting CTHRC1
Chaxiang Li1, He-Sheng Luo1*, Hanqing Hu2, Yufeng Tian2 and Yaoming Zhu2
1Department of Gastroenterology, Renmin Hospital of Wuhan University, Wuhan, PR China
2The First College of Clinical Medical Sciences, China Three Gorges University and Yichang Central People's Hospital, Yichang, Hubei Province, PR China
- *Corresponding Author:
- He-Sheng Luo
Department of Gastroenterology Renmin
Hospital of Wuhan University, PR China
Accepted on March 27, 2017
DOI: 10.4066/biomedicalresearch.29-16-2110
Visit for more related articles at Biomedical ResearchObjective: To investigate the influence of miR-363 on cell proliferation and migration of colorectal cancer.
Methods: The expression of miR-31 in colorectal cancer tissues and colorectal cancer cell lines were measured by RT-PCR. CCK-8 and cell wound scratch assay were used to detect the cell proliferation and migration of colorectal cancer cell lines transfected by miR-363 mimics and miR-363 inhibitor. The expression of CTHRC1 in colorectal cancer cell was detected by Western blotting.
Results: The expression of miR-363 in colorectal cancer cell lines and tissues were obviously up regulated (P<0.05). The cell proliferation and migration ability of colorectal cancer cell was inhibited after transfection of miR-363 mimics. Meanwhile, the expression of CTHRC1 in miR-363 mimics group was (0.31 ± 0.07), which was significantly lower than that in miR-363 inhibition group (2.46 ± 0.37).
Conclusion: miR-363 could inhibit the cell proliferation and migration ability of colorectal cancer cell and the possible mechanism is targeting on CTHRC1.
Keywords
Colorectal cancer, miR-363, CTHRC1, Cell migration.
Background
Colorectal cancer is one of the most common human tumors, the morbidity and mortality of which in the world rank top 3 and top 4, respectively. Moreover, the 5-year survival rate of patients was only about 50% [1]. The occurrence of colorectal cancer is a complex process controlled by multiple genes, and late colorectal cancer patients have poor sensitivity to chemotherapy with poor prognosis. MicroRNAs (miRNAs) are a class of non-coding single-stranded small RNAs of about 22 nucleotides that can inhibit the mRNA translation process by specifically promoting the degradation of certain mRNAs. Thus the regulation of certain target genes was realized, also participating in cell growth, differentiation and apoptosis [2]. At present, 500 kinds of miRNA have been found, of which nearly half of the tumor is closely related to the process [2]. Collagen triple helical repeat protein 1 (CTHRC1) is a secretory glycoprotein selected in the process of arterial injury, with the function of cell migration promotion and collagen deposition reduction [3]. In recent years, CTHRC1 has been found to be abnormally expressed in various solid tumors, and may be involved in the process of tumor invasion and metastasis. In this study, we examined the expression of miR-363 in colorectal cancer tissues and cells, and to investigate its effect on the proliferation and migration of colorectal cancer cells. Besides, bioinformatics software was employed to predict the target gene of miR-363, and its possible mechanism of action was discussed.
Materials and Methods
General conditions
96 patients (mean age 51.9 ± 6.1 years) with colorectal cancer were selected from August, 2013 to October, 2015. All patients were diagnosed by histopathology and had not received any chemotherapy or radiotherapy before operation. The specimens were resected from the colorectal cancer tissues and adjacent normal tissues (>5 cm from the edge of the foci), and then the samples were stored in liquid nitrogen. Six colorectal cancer cell lines (DLD-1, SW480, SW620, Caco-2, HCT116 and HT-29) and normal colorectal epithelial cell FHC were purchased from Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences. Optical quantitative PCR instrument 7500 (the United States ABI), Microplate reader PerkinElmer (US Enspire), Inverted microscope IX51 (Japan Olympus Corporation), RNA extraction kit (Takara, Japan), Lipofectamine 2000 (Invitrogen, U.S.A.), Lipofectamine 2000 (Invitrogen, U.S.A.), an optical quantitative PCR instrument 7500 (ABI, USA), MiR-363 mimics and miR-363 inhibitor (Guangzhou Rui Bo biological), Mouse anti-human CTHRC antibody and GAPDH antibody (Santa Cruz, USA).
Expression of miR-363 in colorectal cancer tissues and cell lines detected by RT-PCR
Total RNA was extracted from fresh frozen tissue and cell lines using RNAisoTM Plus according to the instructions, and CDNA was synthesized using PrimeScriptTM RT Master Mix. The PCR reaction was carried out with the fluorescent quantitative PCR kit and ABI 7500 PCR instrument in strict accordance with the kit instructions. The forward primer sequence of miR-363 was: 5'-CGAATGTCCGTCACATCT-3', and reverse primer sequence 5'-GTGCACGGGCAGAGGT-3'. The PCR conditions were 95°C for 10 min, 95°C for 15 s, 60°C for 60 s, respectively, with a total of 40 cycles. Then the amplication curve was plotted and 2-ΔΔCt analysis was performed (ΔCt for the cycle threshold). The target gene and the internal reference gene CT value were evaluated after comparison of the experimental group and the control group. In other words, ΔΔCt=ΔCt Colorectal cancer tissue -ΔCt normal tissue. All reactions were repeated for 3 times.
Effect of miR-363 on the proliferation of colorectal cancer cells detected by CCK-8
MiR-363 inhibitor, miR-363 inhibitor control were transfected into colorectal carcinoma HCT-116. After transfection 24 h, the cells were seeded in 96-well plates at 1 × 103 (200 μl) for 4 h, following which the cells were adherent. Then 100 μl of culture medium and 10 μl of CCK-8 were added. The cell was placed in CO2 cell incubator at 37°C. CCK-8 kit was used to detect the colorectal cancer cell proliferation ability at different time points (1 d, 2 d, 3 d, 4 d, 5 d), and the cell growth curve was drawn. The procedure was strictly in accordance with the CCK-8 kit instructions.
Transwell method to detect the miR-363 on colorectal cancer cell migration ability
Colon cancer cells SW480 and SW620 in logarithmic growth phase were digested and made into single cell suspensions. The cells were seeded in a 6-well plate, adjusted to a cell density of 2.5 × 105 cells/ml with 1640 medium containing 10% fetal bovine serum. After 24 h, when the cell confluence rate reached 90% or more, the scratch test was performed to detect cell migration ability. Under normal microscope, the images of cell migration were observed and collected at 0 h, 24 h and 48 h after culture. The images were analysed and the mobility was calculated.
Protein expression of CTHRC1 detected by Western blot
The total protein was extracted from the colorectal cancer cells after analysis, and the protein concentration was determined by BCA kit. 12% separation gel and 5% concentrated gel were prepared and loaded at a loading of 40 μg. The samples were transferred to PVDF membrane at 60 V and 100 V, and the PVDF membrane was placed in blocking solution for 1 h at room temperature. CTHRC antibody diluted at 1: 1000 was added and incubated overnight at 4°C. PVDF membrane was washed by TBST for 3 times, adding 1: 5000 washing membrane with peroxidase labeled secondary antibody. The PVDF membrane was incubated at room temperature for 1 h, and the coloration was realized after the X-ray film photosensitive, development and fixing. At last, Quantity One software was used for image analysis.
Statistical analysis
Data were processed by SPSS19.0 statistical software analysis, and all data were represented as mean ± standard deviation (x ± s). Differences between groups were compared using t test or variance analysis. The difference was statistically significant when P<0.05. The miR-363 target gene was predicted by software TargetScan and miRanda.
Results
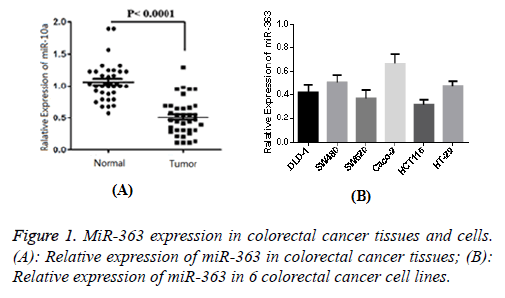
Expression of miR-363 in colorectal cancer tissues and cell lines
The result of 1 case of normal tissue adjacent to the cancer tissue was set as 1, and then the relative expression of miR-363 in 96 cases of cancer was determined as 0.49 ± 0.13. Similarly, the relative expression of adjacent cancer tissues was determined as 1.08 ± 0.24. Figure 1 showed that the difference was statistically significant (P<0.05). The expression level of miR-363 in normal colorectal epithelial cells was 1, and relative expression levels of miR-363 in six colorectal cancer cell lines (DLD-1, SW480, SW620, Caco-2, HCT116 and HT-29) were 0.41 ± 0.06, 0.50 ± 0.07, 0.36 ± 0.06, 0.69 ± 0.09, 0.33 ± 0.05 and 0.49 ± 0.06, respectively. Figure 1B showed that the difference was statistically significant compared with normal cells (P<0.05), indicating that MiR-363 expression in colorectal cancer tissue and cell lines were significantly decreased.
Effect of miR-363 on colorectal cancer cell proliferation activity
After miR-363 inhibitor transfection, the expression of miR-363 in colorectal cancer cell HCT-116 was significantly decreased, while significantly increasing after transfection with miR-363 mimics. The relative expression of miR-363 mimics group was (7.48 ± 0.76), and (0.41 ± 0.08) for miR-363 inhibitor, which showed significant difference compared with blank group (P<0.05). Growth curves of colorectal cancer cells were drawn according to different time points. The viability of cells transfected with miR-363 inhibitor was significantly increased, while significantly lower for transfected miR-363 mimics group (P<0.01).
Effect of miR-363 on migration of SW480 and SW620 cells in colorectal cancer
Cell scratch test showed that migration rates of SW480 and SW620 in colorectal cancer cells were 0.279 ± 0.019 and 0.261 ± 0.019, respectively. The rate was lower than 0.721 ± 0.029 and 0.731 ± 0.032 of transfected blank plasmid, showing the statistical difference (P<0.05).
Effect of miR-363 on CTHRC1 protein expression
The target genes of miR-363 were predicted by TargetScan and miRanda. As a result, miR-363 was found to bind to the 3'UTR end of CTHRC1. To further test the possible effect of miR-363 on CTHRC1 protein, the expression of CTHRC1 protein in colorectal cancer cell line HCT-116 was detected 24 h after transfection. The results showed that the expression level of CTHRC1 in the colorectal cancer cells transfected with miR-363 mimics (0.31 ± 0.07) was significantly lower than that in the control group. While the relative expression of CTHRC1 in transfected miR-363 inhibitor group was (2.46 ± 0.37), which is significantly higher than that in control group (P<0.05). This result indicated that miR-363 could significantly inhibit the expression of CTHRC1 protein.
Discussion
MiRNA is a non-coding RNA of about 22 nt in length. By silencing specific genes, miRNAs can degrade certain mRNAs and thereby inhibit the synthesis of specific proteins. Thus, miRNAs regulated gene expression levels at the transcriptional level [4]. Studies have confirmed that miRNA can affect a variety of physiological and pathological process, such as regulation of cell proliferation and apoptosis. In recent years, the roles of miRNAs in tumorigenesis have attracted increasing attention. Moreover, certain miRNAs were directly involved in the formation of human tumors. Tumor progression was a complex process involving multiple gene regulation and multiple steps, where miRNA expression played an important role. However, the molecular mechanism of the regulation of miRNAs was still unclear.
MiR-363 was a new type of small RNA discovered in recent years. As a member of the miRNA family, it participates in the development and progression of some tumors. MiR-363 was first identified in head and neck tumors with lymph node metastases. It was also down-regulated in some highly invasive tumor cell lines. Further studies have shown that miR-363 can affect the invasion and migration of tumor cells by targeting the PDPN protein [1,5]. Li et al. have shown that miR-363 can regulate the development of neuroblastoma and the process of metastasis [6]. According to the different tumor tissue,miR-363 expression level and its function varied. Sun et al. found that miR-363 had significantly different expression in renal cell carcinoma, gastric cancer, colorectal cancer, head and neck tumor and neuroblastoma tumor tissue [7]. Other studies have shown that miR-363 was related to clinical stage and lymph node metastasis of colorectal cancer patients [8]. However, its specific function was rare reported.
The results of this study showed that miR-363 was significantly lower in colorectal cancer tissues. Similarly, the expression level in six colorectal cancer cell lines was also significantly lower than that in normal colorectal mucosa cells. This indicated that miR-363 was involved in the development and progression of colorectal cancer. In this study, we transfected miR-363 mimics and miR-363 inhibitor to control the level of miR-363 in human colorectal cancer cell line HCT-116. The results of CCK-8 showed that the growth rate of miR-363 transfected cells decreased significantly, while the growth rate of miR-363 inhibitor transfected cells increased significantly. This suggests that miR-363 may play an inhibitory role in the development of colorectal cancer by inhibiting the proliferation of colorectal cancer cells. Furthermore, the migration of colorectal cancer cell SW480 and SW620 after transfection of miR-363 mimics was significantly decreased, suggesting that miR-363 can inhibit the migration of colorectal cancer cells.
The genesis and development of colorectal is regulated by many genes and proteins. The research of these related genes and proteins is of great significance to understand the pathogenesis of colorectal. CTHRC1 is a 25 kD-sized secretory glycoprotein, whose main function is to inhibit the synthesis of collagen mechanisms. CTHRC1 protein are abnormal expressed in malignant tumors, such as breast cancer, colorectal cancer, liver cancer and esophageal cancer [9,10]. Gu et al. found that CTHRC1 expression was significantly reduced in non-invasive melanoma by immunohistochemistry, while in metastatic and invasive melanoma tissues, the expression was significantly up-regulated [11]. Tan et al. also found that the expression of CTHRC1 in colorectal cancer was significantly correlated with the differentiation and clinical stage of colorectal cancer [12]. Oh et al. found that CTHRC1 was highly expressed in colorectal cancer, increasing the cell proliferation, migration and invasive ability. While targeting silencing of CTHRC1 could reduce the proliferation of colorectal cancer cell line HT-29 [13]. The results showed that the expression of CTHRC1 protein was significantly decreased after overexpression of miR-363 in colorectal cancer cell line HCT-116. In contrast, the expression of CTHRC1 was significantly increased after miR-363 inhibition, indicating that miR-363 inhibited the expression of CTHRC1 in colorectal cancer cells.
In summary, miR-363 was lowly expressed in colorectal cancer tissues and cell lines. Therefore, overexpression of miR-363 inhibited the proliferation of colon cancer cells and the migration of tumor cells. Hence, miR-363 may be a potential therapeutic target and diagnostic marker for colorectal cancer.
References
- Luo X, Burwinkel B, Tao S. MicroRNA signatures: novel biomarker for colorectal cancer. Cancer Epidemiol Biomark Prev 2011.
- Yang L, Belaguli N, Berger DH. MicroRNA and colorectal cancer. World J Surg 2009; 33: 638-646.
- Tan F, Liu F, Liu H, Hu Y, Liu D. CTHRC1 is associated with peritoneal carcinomatosis in colorectal cancer: a new predictor for prognosis. Med Oncol 2013; 30: 473.
- Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer 2009; 125: 2737-2743.
- Li X, Zhang G, Li Q. Identification of aberrantly expressed miRNAs in rectal cancer tissue. J Central South Univ 2012; 7: 662-668.
- Li W, Shi X, Lv S. Prediction of miR-363 target genes and bioinformatics analysis. Acta Universitatis Medicinalis Nanjing 2015; 7: 911-915.
- Sun Q, Zhang J, Cao W, Wang X, Xu Q. Dysregulated miR-363 affects head and neck cancer invasion and metastasis by targeting podoplanin. Int J Biochem Cell Biol 2013; 45: 513-520.
- Zhang R, Li Y, Dong X. MiR-363 sensitizes cisplatin-induced apoptosis targeting in Mcl-1 in breast cancer. Med Oncol 2014; 31: 1-10.
- Wang P, Wang Y C, Chen X Y. CTHRC1 is up-regulated by promoter demethylation and transforming growth factor-β1 and may be associated with metastasis in human gastric cancer. Cancer Sci 2012; 103: 1327-1333.
- Ke Z, He W, Lai Y, Guo X, Chen S. Overexpression of collagen triple helix repeat containing 1 (CTHRC1) is associated with tumour aggressiveness and poor prognosis in human non-small cell lung cancer. Oncotarget 2014; 5: 9410-9424.
- Gu L, Liu L, Zhong L, Bai Y, Sui H. Cthrc1 overexpression is an independent prognostic marker in gastric cancer. Hum Pathol 2014; 45: 1031-1038.
- Tan F, Liu F, Liu H, Hu Y, Liu D. CTHRC1 is associated with peritoneal carcinomatosis in colorectal cancer: a new predictor for prognosis. Med Oncol 2013; 30: 473.
- Oh JTK, Kim BY, Lee SJ. Collagen triple helix repeat containing 1 (CTHRC1) acts via ERK-dependent induction of MMP9 to promote invasion of colorectal cancer cells 2014.