ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2017) Volume 28, Issue 12
The impact of ΔmtDNA4977 mutation on ulcerative colitis
1Department of Biology, Science and Research Branch, Islamic Azad University, Tehran, Iran
2Department of Biology, Faculty of Sciences, University of Guilan, Rasht, Iran
3Gastrointestinal and Liver Diseases Research Center, Guilan University of Medical Sciences, Rasht, Iran
- *Corresponding Author:
- Zivar Salehi
Department of Biology, Faculty of Sciences
University of Guilan, Rasht, Iran
Accepted date: July 30, 2016
Ulcerative colitis (UC) is an inflammatory bowel disorder characterized by continuous colonic mucosal inflammation that extends proximally from the rectum. The pathogenic mechanisms underlying UC remain unknown. Free-radical-elicited oxidative damage to macromolecules has been established as a contributory factor in UC. It is well known that the mitochondrial genome is more susceptible to increased oxidative damage than nuclear DNA. One of the best-described mtDNA mutations is the ΔmtDNA4977 (mtDNA 4977 bp) or common deletion. The aim of this study was to investigate the relationship between ΔmtDNA4977 and UC. We studied the ΔmtDNA4977 150 patients with UC (64 men, 86 women; mean age, 36.7 ± 11.6 years) and 250 disease-free controls (137 men, 113 women; mean age, 35 ± 12.1 years) in northern Iran. After DNA extraction from colon biopsies, gap polymerase chain reaction (Gap-PCR) was performed. The frequency of ΔmtDNA4977 was significantly different between the UC patients and the control group (78.7% vs. 16.8%, P<0.001). We found that ΔmtDNA4977 was more likely to be present in the patients of younger age (≤ 50 years, P=0.021). The mtDNA deletion was also significantly associated with the increased risk of an extensive form of colitis. Our study indicates that the ΔmtDNA4977 may play a role in the pathogenesis of UC. However, the findings need to be verified in large population-based prospective studies for more rigorous analyses of subgroups.
Keywords
Ulcerative colitis, ΔmtDNA4977, Mutation, mtDNA deletion.
Introduction
Ulcerative colitis (UC) is a chronic inflammation of the large intestine that, together with Crohn’s disease (CD), makes up the majority of inflammatory bowel diseases (IBD) [1]. The prevalence of UC is relatively static in northern Europe and North America, but it is rising in southern Europe and Asia [2]. The estimated prevalence of UC is 15 per 100000 persons in Iranian population [3]. Although the age of onset of UC typically occurs between the ages of 15-40 years, but the disease can occur at any age from infancy to the elderly. It was reported that almost 5% of new cases occur after age 60 [4]. Both sexes are equally affected.
The etiology of this disease is unknown, however, underlying genetic, environmental and lifestyle issues can affect an individual’s predisposition to this disease [5]. Genetic factors involved in the regulation of the immune system are thought to play a significant role in the pathogenesis of UC [6]. The activated genes involved in mucosal inflammation include cytokines IL-6, IL-8 IL-1β, IL-10, TNF-α and ICAM [7], and increased activation of NF-kB in human UC tissue, NOD2 and Vitamin D receptor [8-11]. Different gene polymorphisms such as CAT, IL-23 receptor and P53 have been shown to be related to UC [12-14].
The human mitochondrial genome comprises of a 16.5-kb circular double-stranded DNA molecule that encodes 13 polypeptides of the respiratory chain, 22 transfer RNAs, and two ribosomal RNAs required for protein synthesis [15]. Mitochondrion is the major source of reactive oxygen species (ROS). Oxidative stress has deleterious effects on cell membrane, DNA and mitochondria. The level of oxidative damage in mtDNA is about 10 to 20 times more rapid than the mutation rate in nucleic DNA (nDNA).
The high mutation rate of mtDNA is due to the lack of protective histone proteins or lack of DNA repair systems [16]. Mitochondrial DNA mutations have been shown to be implied in many cancers and inflammatory diseases [17,18]. So far, over 150 mtDNA mutations have been reported in different diseases [19]. More than 100 different mitochondrial DNA deletions have been identified in patients with mitochondrial dysfunction [20]. The mtDNA mutation including large deletion, missense, frameshift, and small deletions/insertions are associated with various types of diseases [21].
The most frequently reported mtDNA mutations are A3243G, found in patients with mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes (MELAS), A8344G in myoclonus epilepsy (MERRF), T8993G/C in neuropathy, ataxia and retinitis pigmentosa (NARP) and Leigh syndrome, large mtDNA deletion that causes Kearns- Sayre syndrome and ΔmtDNA4977 or common deletion. The position of ΔmtDNA4977 is 8483–13459 that embraces multiple structural genes (ATPase 6/8, COIII, ND3, ND4L and ND4) and six tRNA genes. Defects in the respiratory enzymes, the protein products of these fragments, can cause defect in ATP production [22].
Oxidative stress has been well documented in UC patients with increased ROS levels and decreased antioxidant levels in the inflamed mucosa, which ultimately contribute to chronic tissue damage [23]. UC exhibits deficiencies in antioxidant defences, probably because of excessive inflammation [24]. In this study, the relationship between ΔmtDNA4977and UC was investigated in northern Iran.
Materials and Methods
Subjects
To trace the 4977 bp deletion, 150 patients with UC (64 males and 86 females) and 250 normal controls (137 males and 113 females) were included in this study. The diagnosis of UC was based on standard clinical and endoscopic and histopathologic findings. All the patients were from the North of Iran, and were recruited between July 2013 and April 2015. The location of the disease was defined according to the Montreal classification (E1, Ulcerative proctitis; E2, Left sided UC (distal UC); E3, pancolitis or Extensive UC) and the severity was classified as S0, S1, S2, S3 [25].
The control group composed of age- and gender-matched subjects who had undergone an endoscopic examination in the same recruitment period as the UC patients, without evidence of UC. Both the UC and control group samples were collected after obtaining informed consent. This study was conducted after review and approval of the ethics committee of Science and Research Branch, Azad University, Tehran, Iran. The present study was performed in accordance with the declaration of Helsinki (2013 revision).
Total DNA extraction
For each patient, biopsy samples (50-100 mg) were obtained from inflamed mucosa during colonoscopy and collected in 2 ml tubes. Total DNA was extracted from each sample using GPP Solution Kit (Gene Pajoohan Pouya, Tehran, Iran), conventional proteinase K digestion and incubation at 55°C overnight. DNA purity and concentrations were determined by spectrophotometric measurement of the absorbance at 260 and 280 nm by UV spectrophotometer. The extracted DNA samples was placed into a 1.5 ml micro-centrifugal tube and stored at -80°C.
Polymerase chain reaction
To detect the existence of mtDNA in DNA extracts and ΔmtDNA4977, two primer pairs were used (Cinnagen, Tehran, Iran). The mtDNA genotyping was performed as described previously [26]. The PCR reactions (25 μl) were performed in 0.2 μg DNA templates, 10 pmol of each of the forward and reverse primers, 1 × PCR buffer, 1.5 mmol/L MgCl2, 200 μM of each dNTP and 0.5 unites of Taq DNA polymerase (Fermentas, USA). The PCR conditions included an initial denaturation at 95°C for 5 min, then 35 cycles at 94°C for 30s (denaturation), 61.5°C for 1min (annealing), 72°C for 30 s (extension), and a final extension at 72°C for 10 min. After the reaction, 5 μl of PCR products was mixed with 2.5 μl of 6X loading buffer. The PCR products were electrophresed in 2% agarose gel and were visualized under UV illumination. A random of 20% of the sample was regenotyped by another laboratory member to improve the quality of genotyping and its validity and no discrepancy in analysis was found.
Statistical analysis
Statistical analysis was performed using the χ2 test and the MedCalc v.12.1 software (version 12.1, Belgium). Odds ratios (OR) were calculated together with their 95% confidence intervals (CI). A P-value <0.05 was considered to be statistically significant.
Results
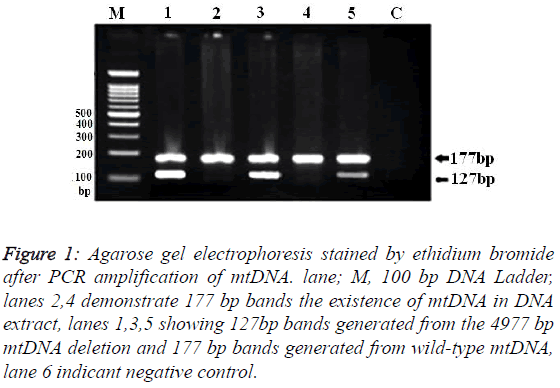
Samples from 150 patients with UC and 250 control subjects were successfully genotyped for the ΔmtDNA4977. Mean ages were 36.7 ± 11.6 years (range: 19- 64) for UC patients and 35 ± 12.1 years (range: 17-67) for the healthy controls. Demographic and clinical characteristics of the UC patients and controls are shown in Table 1. No significant differences were found between the UC cases and controls in age and sex. Of the all patients with UC, 32 (21.3%) had proctitis, 47 (31.3%) had left-sided and 71 (47.3%) had pancolitis. Molecular analysis of the results showed the presence of two separate bands (with lengths of 177-and 127-bp) on the agarose gel (Figure 1).
| Controls (n=250) | UC (n=150) | Characteristics |
|---|---|---|
| Sex | ||
| 137 (54.8%) | 86 (57.3%) | Male |
| 113 (45.2%) | 64 (42.7%) | Female |
| 4.9 (0.2-31) | Duration of disease, mean (years) | |
| 35 ± 12.1 | ||
| 36.7 ± 11.6 | Age of onset (years, mean ± SD) |
|
| Clinical type | ||
| 15 | One episode | |
| 47 | Chronic relapse | |
| 88 | Chronic continuous | |
| Disease extent, n (%) | ||
| 32 (21.3%) | ||
| 47 (31.3%) | E1 | |
| 71 (47.3%) | E2 | |
| E3 | ||
| Disease severity, n (%) | ||
| 8 (5.3%) | ||
| 41 (27.3%) | S0 | |
| 92 (61.3%) | S1 | |
| 45 (18%) | 9 (6%) | S2 |
| 205 (82%) | S3 | |
| 31(21%) | Social status (%) | |
| 119 (80%) | ||
| Rural | ||
| Urban | ||
| 150 | ||
| 76 | Medications | |
| 108 | ||
| Sulfasalazine | ||
| Steroids | ||
| Azathioprine |
Table 1: Demographic and clinical features of UC patients and controls.
Figure 1: Agarose gel electrophoresis stained by ethidium bromide after PCR amplification of mtDNA. lane; M, 100 bp DNA Ladder, lanes 2,4 demonstrate 177 bp bands the existence of mtDNA in DNA extract, lanes 1,3,5 showing 127bp bands generated from the 4977 bp mtDNA deletion and 177 bp bands generated from wild-type mtDNA, lane 6 indicant negative control.
Band with length of 177-bp, confirmed the presence of the mtDNA in all extracts is that by amplification the highly conserved 12s region in mtDNA. Presence of 177-bp band and absence of 127-bp fragment was recorded as wild type mtDNA.
In this study, 118 (78.7%) of patients with clinical UC had ΔmtDNA4977, while 42 (16.8%) of controls had the deletion. Strong association (OR=18.2, 95% CI=10.94-30.48, P<0.0001) between ΔmtDNA4977 and UC was confirmed. It was also found that the prevalence of ΔmtDNA4977 is almost equal in men and women. The associations between ΔmtDNA4977 and clinicopathologic features of UC such as extension and severity were evaluated (Table 2). Carrying a ΔmtDNA4977 was significantly associated with increased risk of severe form of colitis (S2+S3/S0+S1: OR=2.3, 95%CI=1.08-5.29, P=0.03) and extensive form of colitis (E3/E1+E2: OR=3.43, 95% CI=1.42-8.26, P=0.005).
| ∆mtDNA4977 | UC(n=150) | Disease status |
|---|---|---|
| N (%) | ||
| Extent | ||
| 19 (59.3) | 32 | E1 |
| 36 (76.5) | 47 | E2 |
| 63 (88.7) | 71 | E3 |
| Severity | ||
| 4 (50) | 8 | S0 |
| 30 (73.1) | 41 | S1 |
| 78 (84.7) | 92 | S2 |
| 6 (66.6) | 9 | S3 |
Table 2: Association of ΔmtDNA4977with extent and severity of UC.
We also found that, unlike the age-dependent accumulation that was expected, the ΔmtDNA4977was detected more frequently in large intestine of UC patients younger than 50 (44%) as compared to patients over 50 (31%) years. The number of mtDNA deletion was not influenced by the use of corticosteroids, sulfasalazine or azathioprine (P>0.05).
Discussion
In this study, we demonstrated the relationship of ΔmtDNA4977 with UC. There were no reports about the relationship between the ΔmtDNA4977 and UC risk among Iranian. We have shown that patients with UC had a higher frequency of ΔmtDNA4977 in colon biopsy than in normal controls. Of 150 UC cases, 78.7% showed the ΔmtDNA4977in colon biopsies and strong association (OR=18.26, P<0.001, 95% CI=10.94-30.48) between mtDNA deletion and UC was confirmed overall.
UC is a lifelong disease arising from an interaction between genetic and environmental factors, observed predominantly in the developed countries of the world [27]. Oxidative stress has been shown to be implicated in the patients with UC, with an increased production of ROS and decreased antioxidants levels in the inflamed mucosa, which ultimately contribute to chronic tissue damage [23].
Increased free radicals in the cell may lead to lipid and protein modifications, DNA damage, apoptosis or cancerogenic cell transformation [28]. ROS are also a major cause of mtDNA mutations, mtDNA mutations have been implicated in various human diseases including cancer and infections [29]. Among the mtDNA mutations, the ΔmtDNA4977 is one of the most frequent. While the number and types of mtDNA deletions vary greatly in different tissues and organs depending on age and the presence of distinct hereditary or acquired pathological conditions [30]. The ΔmtDNA4977was found to occur between two 13-bp direct repeats at positions 13,447-13,459 and 8,470-8,482 [31]. The genes or parts of the genes encoding for ATPase 8 and 6, COXIII, ND3, ND4 and ND4L and ND5 of the mtDNA removes, resulting in an impairment of the mitochondrial oxidative phosphorylation and ATP production [22].
In humans, the large bowel is a significant source of energy, the first source of energy in the form of ATP is oxidative phosphorylation. This process occurs in mitochondria in which mtDNA encodes for essential subunits of the respiratory chain. When mtDNA deletion occurs in cells, an increased oxidative stress may contribute to accumulation of the deleted mtDNA in colon tissue [30]. Moreover, the ΔmtDNA4977 could lead to energy production catastrophes and abnormal ROS generation.
To date few groups have studied the association of the mtDNA deletions with UC risk, but the results were inconsistent. Fukushima and Fiocchi demonstrated that mtDNA deletions are common in colonic epithelial cells (EC) of the human gastrointestinal tract. They also found that multiple types of mtDNA deletions occur in EC from normal and chronically inflamed colon and that both the type and abundance of mtDNA deletions selectively decrease in actively inflamed UC epithelium [32]. It has been shown that mitochondrial loss precedes the development of dysplasia, and it could be used to detect and potentially predict cancer [33]. The high incidence of mtDNA mutation in colorectal tissues of individuals with UC was shown. It was concluded that the rate of DNA mutation is enhanced in their mucosal cells by oxidative stress caused by chronic inflammation and, hence, malignant transformation occurs more easily than in normal subjects [15].
We also shown that the ΔmtDNA4977was more likely to be present in patients of younger age (≤ 50 years, p=0.027). In a similar study on colorectal cancer, it was shown that the frequency of ΔmtDNA4977of the tumor tissue in younger patients (less than 65 years) was more than older patients (over than 65 years) [34]. However, it has been shown that the ΔmtDNA4977 accumulated in tissues such as brain, skeletal and cardiac muscle increases with age [35]. We also found that the frequency of ΔmtDNA4977in extensive UC (47.3% of patients) is more than other groups (ulcerative proctitis, leftsided). The conflicting results of these reports may be due to the differences in sample sizes, and the impact of other genetic and environmental factors.
The present study has some limitations. First, this study was a single-centre investigation on a relatively small scale. Thus, replication studies with large independent cohorts are warranted. Second, we have analyzed only one mtDNA deletion and in future, exploring other genetic variations in the mtDNA would also be interesting.
It is concluded that ΔmtDNA4977 may play a role in the pathogenesis of UC. However, the findings need to be verified in large population-based prospective studies for more rigorous analyses of subgroups.
Acknowledgments
The authors wish to thank the all people in molecular Genetic Laboratory, Department of Biology, IAU, Tonekabon Branch, Islamic Azad University and Science and Research Branch, Tehran, Iran. We would like to thank Dr. Bahman Sajedi and all patients for their kind's assistance in the collection of samples.
References
- Zheng CQ, Hu GZ, Zeng ZS, Lin LJ, Gu GG. Progress in searching for susceptibility gene for inflammatory bowel disease by positional cloning. World J Gastroenterol 2003; 9: 1646-1656.
- Loftus EV Jr. Clinical epidemiology of inflammatory bowel disease: Incidence, prevalence, and environmental influences. Gastroenterology 2004; 126: 1504-1517.
- Shayesteh AA, Saberifirozi M, Abedian S, Sebghatolahi V. Epidemiological, Demographic, and Colonic Extension ofUlcerative Colitis in Iran: A Systematic Review. Middle East J Dig Dis 2013; 5: 29-36.
- Parray FQ, Wani ML, Malik AA, Wani SN, Bijli AH. Ulcerative colitis: a challenge to surgeons. Int J Prev Med 2012; 3: 749-763.
- Qin X. Etiology of inflammatory bowel disease: a unified hypothesis. World J Gastroenterol 2012; 18: 1708-1722.
- Latiano A, Annese V. Genetics and ulcerative colitis: what are the clinical implications? Curr Drug Targets 2011; 12: 1383-1389.
- Yasukawa K, Tokuda H, Tun X, Utsumi H, Yamada K. The detrimental effect of nitric oxide on tissue is associated with inflammatory events in the vascular endothelium and neutrophils in mice with dextran sodium sulfate-induced colitis. Free Radic Res 2012; 46: 1427-1436.
- Piechota-Polanczyk A, Fichna J. Review article: the role of oxidative stress in pathogenesis and treatment of inflammatory bowel diseases. Naunyn Schmiedebergs Arch Pharmacol 2014; 387: 605-620.
- Gan HT, Chen YQ, Ouyang Q. Sulfasalazine inhibits activation of nuclear factor-kappaB in patients with ulcerative colitis. J Gastroenterol Hepatol 2005; 20: 1016-1024.
- Freire P, Cardoso R, Figueiredo P, Donato MM, Ferreira M. NOD2 gene mutations in ulcerative colitis: useless or misunderstood? Int J Colorectal Dis 2014; 29: 653-661.
- Pei FH, Wang YJ, Gao SL, Liu BR, DU YJ. Vitamin D receptor gene polymorphism and ulcerative colitis susceptibility in Han Chinese. J Dig Dis 2011; 12: 90-98.
- Khodayari S, Salehi Z, Fakhrieh Asl S, Aminian K, Mirzaei Gisomi N. Catalase gene C-262T polymorphism: importance in ulcerative colitis. J Gastroenterol Hepatol 2013; 28: 819-822.
- Hayatbakhsh MM, Zahedi MJ, Shafiepour M, Nikpoor AR, Mohammadi M. IL-23 receptor gene rs7517847 and rs1004819 SNPs in ulcerative colitis. Iran J Immunol 2012; 9: 128-135.
- Vaji S, Salehi Z, Aminian K. Association of p53 codon 72 genetic polymorphism with the risk of ulcerative colitis in northern Iran. Int J Colorectal Dis 2011; 26: 235-238.
- Nishikawa M, Oshitani N, Matsumoto T, Nishigami T, Arakawa T. Accumulation of mitochondrial DNA mutation with colorectal carcinogenesis in ulcerative colitis. Br J Cancer 2005; 93: 331-337.
- Graff C, Bui TH, Larsson NG. Mitochondrial diseases. Best Pract Res Clin Obstet Gynaecol 2002; 16: 715-728.
- Kara M, Tatar A, Borekci B, Dagli F, Oztas S. Mitochondrial DNA 4977 bp Deletion in Chronic Cervicitis and Cervix Cancers. Balkan J Med Genet 2012; 15: 25-29.
- Greaves LC, Taylor RW. Mitochondrial DNA mutations in human disease. IUBMB Life 2006; 58: 143-151.
- Vieira HL, Belzacq AS, Haouzi D, Bernassola F, Cohen I. The adenine nucleotide translocator: a target of nitric oxide, peroxynitrite, and 4-hydroxynonenal. Oncogene 2001; 20: 4305-4316.
- Servidei S. Mitochondrial encephalomyopathies: gene mutation. Neuromuscul Disord 2004; 14: 107-116.
- Modica-Napolitano JS, Singh KK. Mitochondrial dysfunction in cancer. Mitochondrion 2004; 4: 755-762.
- Wallace DC, Lott MT, Lezza AM, Seibel P, Voljavec AS, Shoffner JM. Mitochondrial DNA mutations associated with neuromuscular diseases: analysis and diagnosis using the polymerase chain reaction. Pediatr Res 1990; 28: 525-528.
- Sturniolo GC, Mestriner C, Lecis PE, D'Odorico A, Venturi C. Altered plasma and mucosal concentrations of trace elements and antioxidants in active ulcerative colitis. Scand J Gastroenterol 1998; 33: 644-649.
- Buffinton GD, Doe WF. Depleted mucosal antioxidant defences in inflammatory bowel disease. Free Radic Biol Med 1995; 19: 911-918.
- Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 2006; 55: 749-753.
- Gashti NG, Salehi Z, Madani AH, Dalivandan ST. 4977-bp mitochondrial DNA deletion in infertile patients with varicocele. Andrologia 2014; 46: 258-262.
- Lakatos PL. Recent trends in the epidemiology of inflammatory bowel diseases: up or down? World J Gastroenterol 2006; 12: 6102-6108.
- Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol 2007;39(1):44–48.
- Lu J, Sharma LK, Bai Y. Implications of mitochondrial DNA mutations and mitochondrial dysfunction in tumorigenesis. Cell Res 2009; 19: 802-815.
- Graff C, Clayton DA, Larsson NG. Mitochondrial medicine--recent advances. J Intern Med 1999; 246: 11-23.
- Samuels DC, Schon EA, Chinnery PF. Two direct repeats cause most human mtDNA deletions. Trends Genet 2004; 20: 393-398.
- Fukushima K, Fiocchi C. Paradoxical decrease of mitochondrial DNA deletions in epithelial cells of active ulcerative colitis patients. Am J Physiol Gastrointest Liver Physiol 2004; 286(5):804-13.
- Ussakli CH, Ebaee A, Binkley J, Brentnall TA, Emond MJ. Mitochondria and tumor progression in ulcerative colitis. J Natl Cancer Inst 2013; 105: 1239-1248.
- Chen T, He J, Shen L. The mitochondrial DNA 4,977-bp deletion and its implication in copy number alteration in colorectal cancer. BMC Medical Genetics 2011; 12:18.
- Mohamed SA, Wesch D, Blumenthal A, Bruse P, Windler K. Detection of the 4977 bp deletion of mitochondrial DNA in different human blood cells. Exp Gerontol 2004; 39: 181-188.