ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2019) Volume 30, Issue 2
Survey of ermA, ermB, ermC and mecA genes among Staphylococcus aureus isolates isolated from patients admitted to hospitals in Tehran, Iran by PCR and Sequencing
Ghazaleh Talebi, Ali Hashemia, Hossein Goudarzi*, Aref Shariati, Narjess Bostanghadiri, Javad Yasbolaghi Sharahi and Elham Abbsi
Department of Microbiology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- *Corresponding Author:
- Hossein Goudarzi
Department of Microbiology
School of Medicine
Shahid Beheshti University of Medical Sciences
Tehran, Iran
Accepted date: January 2, 2019
DOI: 10.35841/biomedicalresearch.30-18-1152
Visit for more related articles at Biomedical ResearchStaphylococcus aureus is one of the gram positive bacteria that has created many problems in treatment. The antibiotic resistance is an important problem and debatable topic in the world. In recent years, because of overuse of antibiotics and transition of resistance genes, frequency of resistant staphylococcal infections, are increasing. Clindamycin inductive resistance causes failure in treatment. The aim of current study is detection of clindamycin inductive resistance S. aureus isolates among patients admitted to Tehran hospitals by multiplex PCR. A total of 80 isolates of S. aureus were collected from hospitalized patients in Tehran. The antibiotic susceptibility tests were applied by MIC and disk diffusion methods. The identification of clindamycin inductive resistance isolates was performed by D-zone test. To detection of ermA, ermB, ermC and mecA genes, multiplex PCR was administrated. In current experiment, among 80 isolates, resistance rate to erythromycin and clindamycin were 70% and 45% respectively. By D-zone test, 15 samples were positive. The frequency of ermA, ermB and ermC genes in S. aureus isolates were 5%, 7.5% and 10% respectively. The results of this study demonstrated that the antibiotic resistance is a main problem in patient’s treatment. By identification of resistant isolates and apply appropriate treatment, can be somewhat prevent from outspread of resistant isolates.
Keywords
Methicillin resistance Staphylococcus aureus (MRSA), Clindamycin inductive resistance, D-zone test
Introduction
Staphylococcus aureus species are known as human pathogens which cause skin and soft tissue infections, acute septicemia, pneumonia and toxic shock syndrome [1,2]. These organisms are resistant to most of drugs and constant against most of disinfectants agents [3]. Nowadays, antibiotic resistance of S. aureus is a major problem in society [4]. Different types of antibiotic resistance mechanisms have been found in S. aureus that beta-lactamase producing is the most common which associated gene with enzyme production is located on the plasmid. Resistance to nafcillin by mecA gene which is located on chromosome, resistance to vancomycin (vancomycin and nafcillin resistance genes exist in these strains) and plasmid resistance to tetracycline, erythromycin and aminoglycosides are the types of resistance mechanisms [5,6]. Resistance to nafcillin, methicillin and oxacillin is independent from betalactamase producing [7,8]. The methicillin resistant S. aureus (MRSA) is a problem in development of hospital infections. A study has shown that 38.6% of S. aureus which isolated from hospitalized patients in Shariati hospital and Tehran Children’s Medical Center are MRSA isolates [9]. MRSA are resistant to certain types of oxacillin antibiotics (nafcillin, methicillin, oxacillin and cloxacillin) and all of the beta lactam antibiotics such as penicillin, amoxicillin and cephalosporin’s [10]. The Staphylococcus which are resistant to erythromycin show cross-resistance with macrolides (spiramycin, azithromycin and clarithromycin), lincosamides (clindamycin and lincomycin) and type B streptogramin. Increasing of MLS (Macrolide-Lincosamide-Streptogramin) resistant strains in clinical specimens, indicates increasing of clindamycin utilization [11]. Erythromycin and clindamycin are different classes of antibiotics which bind to 50S ribosomal subunit of bacteria and inhibit protein synthesis. In Staphylococcus resistance to these antibiotics is create by methylation of target site on ribosome that mostly related to methylase gene erm (rRNA) [12]. The ermA, ermB and ermC are types of erm gene which discovered in Staphylococcus [12,13]. The ermA as a chromosomal gene is the most common and ermB which is located on plasmid have not found in primates. The frequency of these plasmids (85-98%) increases by use of erythromycin and clindamycin. 95% of acne causing S. aureus isolates which are under influence of clindamycin have ermC gene [4]. Also, resistance to macrolides can occur through efflux which is dependent to mrsA gene, the other resistance mechanism is inactivation of lincosamides by chemical alterations that related to inuA gene that rarely occurred [12,13]. The routine antibiotic susceptibility tests for S. aureus with inducible resistance, demonstrate the resistance to erythromycin and susceptibility to clindamycin. Thus, clindamaycin therapy may be given for erm and mrsA mutants, which ultimately leads to failure in therapy [12]. On the other side, clindamycin is one of the most commonly used antibiotics, especially for treatment of skin and soft tissue infections caused by S. aureus and is a supersede drug in patients with allergies to penicillin [14]. Induced resistance to clindamycin in S. aureus is common which is not detected by routine tests and according to CLSI (Clinical and Laboratory Standards Institute) guidelines, Dzone test is the best method to identity of clindamycin induction resistance [4,15]. For detection of inducible clindamycin resistance, the standard test is D-zone test. In routine antimicrobial susceptibility tests, those which are clindamycin inducible resistance, emerge as erythromycin resistant and clindamycin susceptible [16]. Resistance rate to erythromycin class is so fast, so these medications should not be used alone for the treatment of chronic infections [17]. This test as a simple and reliable test with high sensitivity and specificity, detects clindamycin inductive resistance in clinical laboratories [2]. Placing erythromycin and clindamycin disks at a distance of 15-20 mm from each other and formation of a flat region close to erythromycin disk demonstrates clindamycin inductive resistance which indicates the presence of erm gene [17]. Therefore, providing a useful solution to use the appropriate treatment regimen by determination of the antibiotic resistance pattern in the target population is necessary. The D-zone test can be an acceptable method to identification of clindamycin inductive resistance of S. aureus isolates. The aim of current study is detection of inducible clindamycin resistance S. aureus isolates among patients admitted to Tehran hospitals by multiplex PCR. Also due to cheapness, being simple and importance of inductive resistance detection in effective treatment of S. aureus, is recommended.
Materials and Methods
In this descriptive study, by referring to the clinical laboratories of the hospitals affiliated to the Shahid Beheshti University of Medical Sciences, the Staphylococcus isolates were collected and transmitted to the Microbiology Lab in Shahid Beheshti University of Medical Sciences. To confirm S. aureus isolates, gram stain, β-hemolysis on blood agar, growth on mannitol salt agar, catalase, coagulase and DNase tests were applied [18]. According to CLSI guidelines (2015), susceptibility of the isolates to erythromycin, clindamycin, oxacillin, trimethoprim, sulfamethoxazole and tetracycline (Mast Co. UK) with disk diffusion method on Mueller Hinton agar (Merck Co. Germany) was performed. S. aureus ATCC25923 was used as a quality control [19]. Determination of MIC (Minimum Inhibitory Concentration) for vancomycin was performed based on CLSI guidelines. S. aureus suspension in BHI (Brain Heart Infusion) broth with 0.5 McFarland concentration was prepared and inoculated on Mueller Hinton agar (Merck Co. Germany). The clindamycin 2 μg and erythromycin 15 μg disks, were placed at a distance of 12 mm from each other and after incubation for 18-24 h in 35°C, preventive zone diameter was measured. D-shaped zone of clindamycin from the side of erythromycin disk was carefully investigated. Extraction of DNA was performed by GeNet Kit (Cat. No. K-3000). To investigate the quality of extracted DNA, in terms of quantity all samples were checked by WPA Bio-wave II Nano spectrophotometer at 260/280 wavelength. Multiplex PCR was performed for ermA, ermB and ermC genes by master mix 2X (Sina Colon Co. Cat. No. PR901638) which included the following instances: 0.4 mmol of each dNTP, 3 mmol MgCl2, 0.08 U/μl Taq-polymerase. The primer sequences for ermA, ermB and ermC genes were as follows respectively: ermA F: AGCGGTAAACCCCTCTGA, ermA R: TTCGCAAATCCCTTCTCAAC with, ermB F: CCGTTTACGAAATTGGAACA GGTAAAGGGC, ermB R: GAATCGAGACTTGAGTGTGC and ermC F: AATTGGCTCAGGAAAAGGGC, ermC R: AAATCGTCAATTCCCGCATG. The PCR product size for ermA, ermB and ermC genes was 190, 360 and 564 bp respectively. The primer sequences were checked by Gene Bank (Blast) and finally the PCR program were as follows: Initial denaturation at 94°C for 5 min for one cycle, secondary denaturation at 94°C for 45 s, the annealing at 58°C for 45 s, the extension at 72°C for 45 s for 36 cycles and final extension at 72°C for 5 min for one cycle. The PCR products were analysed by electrophoresis on 1.5% agarose gel in TBE (Tris- Borate-EDTA), then the gel was stained by ethidium bromide (10.5 μg/ml) afterward the results were checked by Gel Doc in the wavelength of 280 nm.
Results
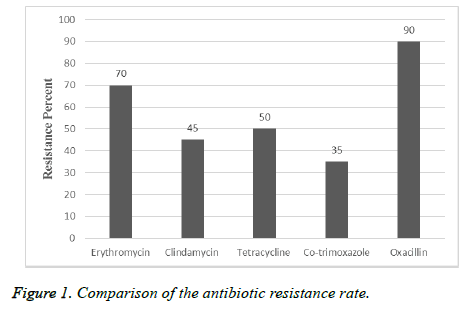
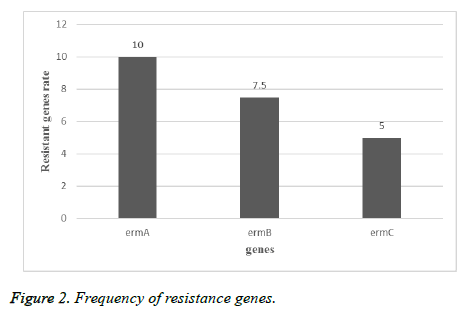
In this descriptive study, from October 2015 to November 2016, samples collection was performed from patients referred to Tehran hospitals. After identification by biochemical and microbial tests, 80 isolates were approved as S. aureus. 75% of samples belong to males and 25% of them referred to females. 45 of isolates were collected from urine, 10 isolates from wound, 5 isolates from tracheal, 4 isolates from sputum, 10 isolates from blood and 6 isolates from other clinical specimens were collected (Table 1). Among 80 isolates of S. aureus the resistance rate to erythromycin was 70%, to clindamycin was 45%, to tetracycline was 50%, to cotrimoxazole was 35% and 90% were resistant to oxacillin (Figure 1). By MIC and disk diffusion results, all of the isolates were susceptible to vancomycin (Figure 2). The Dzone test results demonstrated 15 isolates were positive phenotypically (Figure 3). Among these isolates 55 of them were MRSA (including mecA gene by PCR) and 45 isolates were methicillin susceptible S. aureus (MSSA) which confirmed by cefoxitin disk diffusion agar. The ermA, ermB and ermC genes were detected in 8 (10%), 6 (7.5%) and 4 (5%) of isolates respectively. Also, 2 isolates had all three genes simultaneously.
| Origin of samples | Number of samples |
|---|---|
| Urine | 45 |
| Wound | 10 |
| Blood | 10 |
| Tracheal | 5 |
| Sputum | 4 |
| Other clinical specimens | 6 |
| Total | 80 |
Table 1. Frequency of collected samples.
Discussion
According to the results of other studies in recent years, S. aureus is one of the most common gram positive bacteria which is involved in nosocomial infections. Treatment of infections caused by S. aureus is difficult and causes mortality in hospitalized patients. Due to ability of S. aureus isolates to acquire resistance and overuse of antibiotics, resistance rate is increasing in this bacteria. In current study, 70% of isolates were resistant to erythromycin, 45% to clindamycin, 50% to tetracycline, 35% to co-trimoxazole and 90% to oxacillin and D-zone test of 15 isolates were positive. According to study by Sasirekha et al. in India, 153 isolates of S. aureus were collected and antibiotic susceptibility tests for cefoxitin, oxacillin and D-test to determination of clindamycin inductive resistance were performed. The results showed that 42 (45.27%) of isolates were resistant to methicillin, 43 of isolates were susceptible to clindamycin and 63 (17.41%) of isolates were resistant to erythromycin which 14 of them (15.9%) showed inductive resistance, 29 (95.18%) of isolates had real sensitivity to clindamycin and 20 (13%) of isolates showed intrinsic resistance to clindamycin [20]. The rate of resistance in this study was less than current experiment that reason of this difference can be sample size, the type of antibiotic used in that area or difference in year of sampling. As regards the rate of antibiotic resistance is increasing every year, in a study by Ansari et al. in Nepal, 15718 clinical specimens from blood, urine, sputum and pus were collected, the antibiotic susceptibility tests and identification of resistant isolates to methicillin by cefoxitin, oxacillin disks and identity of inductive resistance to clindamycin by D-zone test, showed 306 of isolates were S. aureus and all of the isolates were susceptible to vancomycin and teicoplanin. Resistance to methicillin by cefoxitin disk in 43.1% and resistance to oxacillin in 39.2% of isolates were observed which 100 of isolates were resistant to erythromycin and inductive resistance of clindamycin was reported in 38 (12.4%) of isolates [21]. The rate of resistance to erythromycin was more than current study that it depends on antibiotic prescribing in those regions, but the rate of resistance to clindamycin were consist with current study. In other study by Chaturvedi et al. in India in, 6468 different clinical specimens were collected and resistance to mupirocin by E-test and agar dilution methods and inductive resistance to clindamycin by D-test were determined. The results of this study showed that 82 of isolates were resistant to methicillin and 15 (18.3%) of isolates were resistant to mupirocin that 8 isolates (53.3%) had high level resistance and 7 isolates (46.7%) had low level resistance and 4 isolates among resistant isolates to methicillin and mupirocin had inductive resistance to clindamycin [22]. Also in other study by Juayang et al. in Philippines, 94 infections caused by S. aureus were detected during 2010-2012 that 38 (40.6%) had methicillin resistance and 37 (39.4%) showed clindamycin inductive resistance. The most samples (71.05%) were wound and abscess specimens with methicillin resistance, while in blood specimens 5.3% was reported. The results of antibiotic susceptibility tests showed that all of the isolates (100%) to linezolid, 95% to tetracycline and 0% to penicillin G were susceptible [23], that the rate of resistance to clindamycin was somewhat similar to the current study. Iraj et al. in Hamedan, Iran, 520 nasal swabs from children under the age of 12 were collected and antibiotic susceptibility tests by disk diffusion (for oxacillin, erythromycin, clindamycin, cefazolin, vancomycin and D-test) showed that 118 (22.3%) of children had S. aureus and just one isolate showed methicillin resistance. Also 5% of these isolates acquired from community, 6.3% of them acquired from hospital and had clindamycin inductive resistance [24] that the rate of resistance was less than present study. The reason of this different can be attributed to the surveyed community, because resistance rate is lower in kids. In a study by Fatemeh et al. in Isfahan, Iran, 585 isolates of S. aureus were collected from 3 clinical centers in Tehran (2005-2006), antibiotic susceptibility tests by disk diffusion for 13 antibiotics, MIC test for methicillin (according to broth micro dilution and PCR) to detection of mecA gene was performed, which in total 321 (54.7%) isolates identified as S. aureus and were resistant to kanamycin (88%), cefotaxime (65%), methicillin (66%), oxacillin (88%), ampicillin (100%), erythromycin (41%), clindamycin (38%), sulfamethoxazole-trimethoprim (41%), vancomycin (0%),chloramphenicol (40%), ciprofloxacin (93%), gentamycin (20%) and tetracycline (64%). All of the isolates were resistant to methicillin and 63% of isolates with interstitial resistance had mecA gene [25]. The resistance rate to erythromycin and clindamycin in this study was less than current study which is related to the time of sampling that by passing the time, due to acquire of resistance elements to antibiotic, resistance of isolates has increased. Also in Razavi et al. experiment in Karaj, Iran, 130 isolates of S. aureus were collected from blood, tracheal aspirate, urine and wound specimens and antibiotic susceptibility test was performed by disk diffusion method also presence of mecA gene was detected by molecular tests. They showed that 100 of isolates were S. aureus and 56% of them had mecA gene which indicates the high prevalence of methicillin resistance in Iran, also 45.4% of methicillin resistant isolates at least were resistant to three classes of antibiotic and just 4 isolates among 130 isolates were susceptible to all of the tested antibiotics and all of the S. aureus isolates were susceptible to vancomycin and resistant to penicillin, 58% to methicillin, 44% to erythromycin, 60% to gentamycin, 56% to ciprofloxacin, 42% to ceftriaxone, 16% to rifampin and 53% to amikacin [26]. Sample type, sampling time, type of the disk which used and sample size are instances that cause alterations in antibiotics percentage. In current experiment among 80 isolates ermA, ermB and ermC genes were detected in 10%, 7.5% and 5% of isolates respectively. In other study by Ghanbari et al. in Isfahan, they were found that 4 isolates had ermC, 2 isolates had ermB and 1 isolate had ermA [26], that current study was similar to this experiment. Also in a study by Mousavian et al. in Ahwaz, 41.1% of isolates had ermA, 17.7% had ermC and none of the isolates had ermB gene [28,29]. The prevalence of some of these genes in this study was much more than current experiment which the reason of these differences can be the region and time of sampling.
Conclusion
The prevalence of antibiotic resistance in current study has been of concern, therefore control of infections, preventing from dissemination of resistant bacteria, need to have accurate management in drug prescription and identification of resistant isolates. Also due to cheapness, being simple and importance of inductive resistance detection in effective treatment of S. aureus, it’s recommended that this test be performed in laboratory routinely that will prevent from reporting of resistant isolates instead of susceptible isolates.
Acknowledgment
This study was funded by Research Department of the School of Medicine, Shahid Beheshti University of Medical Sciences (grant No 6049).
Ethical Statement
The present study was acknowledged by the Ethics Committee of Shahid Beheshti University of Medical Sciences with reference number IR.SBMU.MSP.REC.1396.378.
References
- Goudarzi G, Tahmasbi F, Anbari K, Ghafarzadeh M. Distribution of genes encoding resistance to macrolides among staphylococci isolated from the nasal cavity of hospital employees in Khorramabad, Iran. Iran Red Crescent Med J 2016; 18: 25701.
- Vandana K, Singh J, Chiranjay M, Bairy I. Inducible clindamycin resistance in Staphylococcus aureus: Reason for treatment failure. J Glob Infect Dis 2009; 1: 76-77.
- Dupieux C, Bouchiat C, Larsen AR, Pichon B, Holmes M, Teale C. Detection of mecC-positive Staphylococcus aureus: What to expect from immunological tests targeting PBP2a? J Clin Microbiol 2017; 55: 1961-1963.
- Prabhu K, Rao S, Rao V. Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. J Lab Physicians 2011; 3: 25-27.
- Hiramatsu K, Cui L, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol 2001; 9: 486-493.
- Hiramatsu K. Vancomycin resistance in staphylococci. Clin Microbiol Rev 2002; 15: 430-438.
- Sun HD, Teng SW, Huang BS, Hsiao SM, Yen MS, Peter Wang PH. Combination of ultrasound-guided drainage and antibiotics therapy provides a cosmetic advantage for women with methicillin-resistant Staphylococcus aureus breast abscess. Taiwan J Obstetr Gynecol 2014; 53: 115-117.
- Gordon A, Marshall J, Ramdass K, Stewart-Johnson A, Adesiyun A. Frequency of resistance to methicillin and other antimicrobial agents among Staphylococcus aureus strains isolated from pigs and their human handlers in Trinidad. Infect Ecol Epidemiol 2014; 4.
- Xu Z, Li L, Chu J, Peters BM, Harris ML, Li B. Development and application of loop-mediated isothermal amplification assays on rapid detection of various types of staphylococci strains. Food Res Int 2012; 47: 166-173.
- Zhao X, Forghani F, Park MS, Oh DH. Rapid detection of viable Escherichia coli O157 by coupling propidium monoazide with loop-mediated isothermal amplification. J Microbiol Biotechnol 2013; 23: 1708-1716.
- Schreckenberger PC, Ilendo E, Ristow KL. Incidence of constitutive and inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci in a community and a tertiary care hospital. J Clin Microbiol 2004; 42: 2777-2779.
- Steward CD, Raney PM, Morrell AK, Williams PP, McDougal LK, Jevitt L, McGowan JE Jr, Tenover FC. Testing for induction of clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus. J Clin Microbiol 2005; 43: 1716-1721.
- Khan SA, Nawaz MS, Khan AA, Cerniglia CE. Simultaneous detection of erythromycin-resistant methylase genes ermA and ermC from Staphylococcus spp. by multiplex-PCR. Mol Cell Probes 1999; 13: 381-387.
- Ajantha G, Kulkarni RD, Shetty J, Shubhada C, Jain P. Phenotypic detection of inducible clindamycin resistance among Staphylococcus aureus isolates by using the lower limit of recommended inter-disk distance. Indian J Pathol Microbiol 2008; 51: 376-378.
- Gardiner BJ, Grayson ML, Wood GM. Inducible resistance to clindamycin in Staphylococcus aureus: validation of Vitek-2 against CLSI D-test. Pathology 2013; 45: 181-184.
- Yusuf E, de Bel A, Bouasse J, Piérard D. D-Zone test for detection of inducible clindamycin resistance using SirScan paper disks and Rosco Neo-Sensitabs at 25 and 15 mm distances. J Med Microbiol 2014; 63: 1052-1054.
- Levin TP, Suh B, Axelrod P, Truant AL, Fekete T. Potential clindamycin resistance in clindamycin-susceptible, erythromycin-resistant Staphylococcus aureus: report of a clinical failure. Antimicrob Agents Chemother 2005; 49: 1222-1224.
- Tille P. Bailey and Scotts Diagnostic Microbiology-E-Book: Elsevier Health Sciences 2015; 344-345.
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing? Twenty-Six informational supplement. Document M100-S25 CLSI 2015.
- Sasirekha B, Usha M, Amruta J, Ankit S, Brinda N, Divya R. Incidence of constitutive and inducible clindamycin resistance among hospital-associated Staphylococcus aureus. Biotech 2014; 4: 85-89.
- Ansari S, Nepal HP, Gautam R, Rayamajhi N, Shrestha S, Upadhyay G, Acharya A, Chapagain ML. Threat of drug resistant Staphylococcus aureus to health in Nepal. BMC Infect Dis 2014; 14: 157.
- Chaturvedi P, Singh AK, Singh AK, Shukla S, Agarwal L. Prevalence of mupirocin resistant Staphylococcus aureus isolates among patients admitted to a tertiary care hospital. N Am J Med Sci 2014; 6: 403-407.
- Juayang AC, de los Reyes GB, de la Rama AJG, Gallega CT. Antibiotic resistance profiling of Staphylococcus aureus isolated from clinical specimens in a tertiary hospital from 2010 to 2012. Interdiscip Perspect Infect Dis 2014; 2014.
- Sedighi I, Mashouf RY, Pak N, Rabiee M-AS. D-test method for detection of inducible clindamycin resistance in Staphylococcus aureus. Iran J Pediatr 2009; 19: 293-297.
- Rahimi F, Bouzari M, Maleki Z, Rahimi F. Antibiotic susceptibility pattern among Staphylococcus spp. with emphasis on detection of mecA gene in methicillin resistant Staphylococcus aureus isolates. Arch Clin Infect Dis 2009; 4: 143-150.
- Davoodi NR, Jalil V, Harz N, Hajrafi A, Rajaei B, Gerayesh-Nejad S. Molecular detection of methicillin resistant Staphylococcus aureus (MRSA) and methicillin resistant coagulase-negative Staphylococcus (CoNS) in Iran. Afr J Microbiol Res 2012; 6: 3716.
- Ghanbari F, Ghajavand H, Havaei R, Jami MS, Khademi F, Heydari L. Distribution of erm genes among Staphylococcus aureus isolates with inducible resistance to clindamycin in Isfahan, Iran. Adv Biomed Res 2016; 5: 62.
- Moosavian M, Shoja S, Rostami S, Torabipour M, Farshadzadeh Z. Inducible clindamycin resistance in clinical isolates of Staphylococcus aureus due to erm genes, Iran. Iran J Med Microbiol 2014; 6: 421.
- Gholipourmalekabadi M, Sameni M, Hashemi A, Zamani F, Rostami A, Mozafari M. Silver-and fluoride-containing mesoporous bioactive glasses versus commonly used antibiotics: activity against multidrug-resistant bacterial strains isolated from patients with burns. Burns 2016; 42: 131-140.