ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2008) Volume 19, Issue 2
Quantitative and qualitative coding of Lactate dehydrogenase isoenzymes and their genes: Evolutionary implication in genus Channa (Channidae: Channiformes)
Section of Genetics, Department of Zoology, Faculty of Life Sciences, Aligarh Muslim University, Aligarh, U.P., India
- *Corresponding Author:
- Riaz Ahmad
Section of Genetics
Department of Zoology
Faculty of Life Sciences
Aligarh Muslim University
Aligarh- 202001. (U.P)
India
Mob: 00919927018812
E-mail: riazzool@rediffmail.com
Accepted date: April 07 2008
Lactate dehydrogenase (LDH: 3.1.1.27) is a tetrameric enzyme participates in carbohydrate metabolism by catalyzing the oxidation of lactate and reduction of pyruvate. The random tetramerization of LDH-A and -B subunits leads to the formation of five tetramers: LDH-B4, LDH-A1B3, LDH-A2B2, LDH-A3B1 and LDH-A4. Due to the biochemical and evolution-ary significance of LDH isoenzyme characters or LDH genes, efforts are made here to pro-pose a numerical coding method of quantifying the isoenzyme activities and demonstrate the presence of LDH loci or the changing levels of isoenzymes in different tissues of selected air-breathing teleosts Channa punctata, C. gachua and C. striatus. Polyacrylamide gel electro-phoretic (PAGE) profiles of LDH isoenzymes were utilized and software analysis of gel scans was performed. Our results show a great variation in the quantitative as well as quali-tative presence of isoenzymes or their genes among the selected freshwater congeneric spe-cies. Based on the obtained results, adaptive strategies of Channa species are discussed in re-lation to their evolutionary set up.
Keywords
LDH isoenzymes; differential gene expression; steady-state patterns; polyacrylamide gel electro-phoresis; Channa; evolutionary genetics
Introduction
Lactate dehydrogenase (LDH: 3.1.1.27) enzyme in fishes have always been the subject of much attention [1-4]. But, scarce reports are available from India dealing either with the expression of LDH genes /presence of loci or their quantitative changes in freshwater teleosts [5-7]. Lactate dehydrogenase is one of the chief enzyme of carbohydrate metabolism which catalyses the oxidation of lactate and reduction of pyruvate during anaerobic glycolysis. Usu-ally two subunits -A and -B are present in different tissues as per their metabolic demands. Their random tetrameri-zation results in the formation of two homotetramers: LDH-A4 (muscle specific) and -B4 (heart specific) and, three heterotetramers -A1B3, -A2B2, and -A3B1 [2,8].
LDH genes in fish, LDH-A and LDH-B have been shown homologous with those of higher vertebrates and accord-ing to available reports gene duplication of a single ances-tral form gave rise to these forms nearly five hundred mil-lion years ago, which is also the expected time of origin of fishes [1,9-10]. A third gene LDH-C, which is pre-sumed to be the result of LDH-B gene duplication also shows an orthologous expression and has been demonstrated in the eye or liver of many teleosts [1,3,8].
It is evident that the LDH isoenzymes have also been util-ized as an indispensable tool in systematic and phyloge-netic studies due to either the quantitative/ qualitative dif-ferences or their differential expression [11-12]. There-fore, the significance of these important characters in evo-lutionary protocols cannot be ignored. In the present study, the presence/ absence of LDH isoenzymes or genes and their quantitative differences have been assigned Arabic numerals and utilized to understand the evolution-ary genetics of some selected species of genus Channa: C. punctata, C. gachua and C. striatus. Channids, besides having accessory air-breathing organs, are of special in-terest due to their distinct adaptive lineage or evolutionary group.
Materials and Methods
Procurement of fish
Mature and live fish representing the mixed catch were brought to the laboratory from the fish markets of Aligarh and Bareilly. They were identified and acclimatized in well-aerated water tanks prior further experimentation.
Preparation of tissue extract:
Following the sacrifice of C. punctata, C. gachua and C. striatus, their tissues namely brain, eye, heart, liver, skele-tal muscle and kidneys were carefully dissected and ho-mogenized in chilled 50 mM Tris–HCl buffer of pH, 7.5 as described earlier [13]. The extract was centrifuged at 4 °C and 15000 rpm for 20 min. The clear supernatant ob-tained after centrifugation was used for analysis by poly-acrylamide gel electrophoresis (PAGE).
Protein estimation:
Standard method of Lowry et al. [14] was used for esti-mation of protein concentration in the supernatant of dif-ferent tissues extract of selected fish species.
Electrophoresis and visualization of LDH isoenzyme bands:
Electrophoresis was essentially carried out according to the protocol outlined previously [13]. Initially, the runs were made in 1× upper gel buffer and later replaced with Tris–glycine buffer as the samples entered the lower sepa-rating gel.
LDH isoenzyme bands were stained and visualized in the presence of L-lactate as the substrate according to the procedure of Shaw and Prasad [15]. Then the stained gels were fixed in 7% acetic acid (v/v) and documented via scanning on HP Deskjet F370 All-in-One computer as-sembly.
Quantitative/ qualitative analysis of LDH isoenzymes using softwares:
Quantitative estimates of LDH isoenzymes in the gel-scans was carried out using GelPro software (Media cy-bernetics, USA), while their qualitative coding were made by Scion Imaging software (Beta release-4, Scion Corpo-ration) respectively. Arabic numerals (0-4) were assigned to each isoenzyme activity as per the band peak or peak area corresponding to each LDH tetramer. But, while per-forming qualitative analysis numerals 1 and 0 were cho-sen to represent the presence or absence of a specific LDH gene/loci.
Results
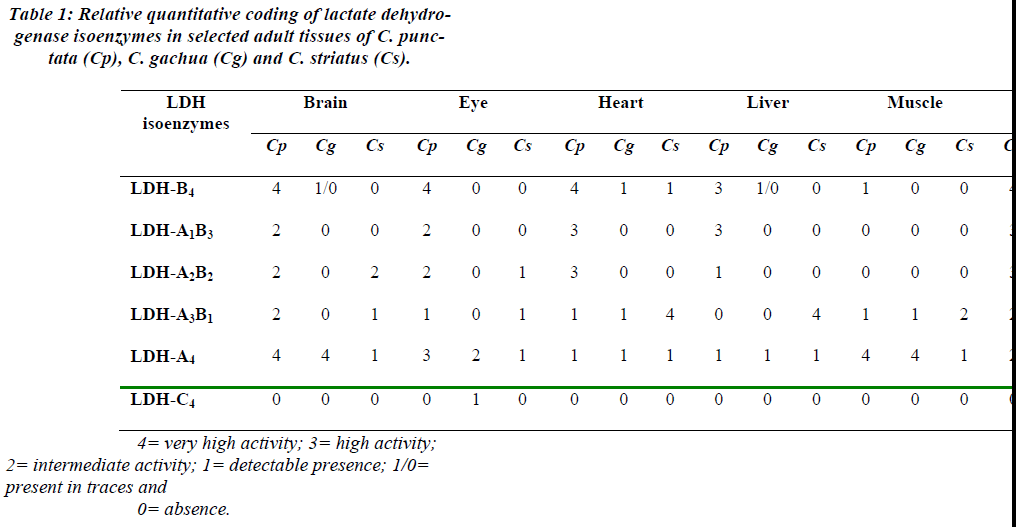
In the investigated teleosts, typical zymograms show de-creasing electrophoretic mobility from anode to cathode in the order of LDH-A4, LDH-A3B1, LDH-A2B2, LDH-A1B3 and LDH-B4. Since during the present study six tissues namely brain, eye, heart, liver, skeletal muscle and kidneys were selected as the source of examining the LDH isoenzyme activities, on individual tissue basis their coding in Arabic numerals was performed (Table-1). It was observed that brain of C. punctata possessed very high activities of homotetramers LDH-B4 and -A4. While C. gachua demonstrated LDH-A4 as the band of very high activity and C. striatus showed the detectable presence of it. In the eye of C. punctata LDH-B4 predominated whereas in case of C. gachua, LDH-A4 was detected as the major activity along with the detectable presence of LDH-C4. This LDH-C4 isoenzyme was unambiguously detected in the entire collection of C. gachua from local fish market of Aligarh or Bareilly district. Heart specific LDH-B4 was observed as a band of very high activity in C. punctata while in other two sister species it was in de-tectable amounts. In the heart of C. striatus instead of -B4, LDH-A3B1 was observed as the major activity.
Liver of C. punctata shows high activity of LDH-B4 which was almost absent in C. striatus and detected as trace activity in C. gachua. Very high activity of LDH-A4 was there in skeletal muscle of C. punctata as well as C. gachua. LDH-B4 was the main activity in the kidneys of C. punctata while other two species were lacking it. Activity of LDH-A3B1 in C. striatus vary from very high to intermediate to again very high in liver, skeletal muscle and kidneys respectively (Table 1).
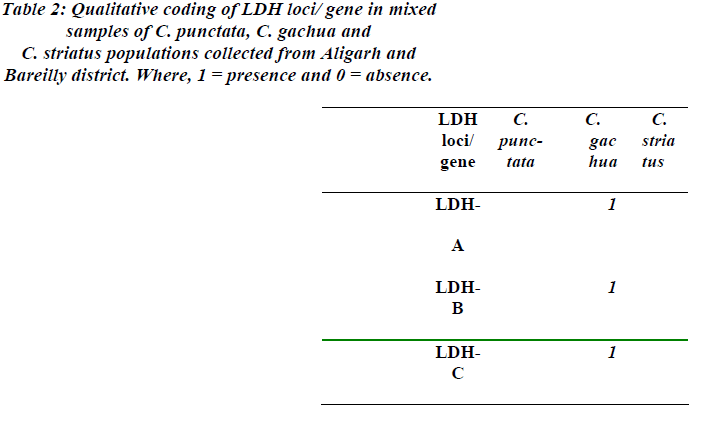
The present results also demonstrate the presence of three LDH loci in the investigated populations of selected teleosts. In the populations of C. punctata, C. gachua and C. striatus from Aligarh and Bareilly, consistent expres-sion of LDH-A and -B genes was recorded. In addition to it, LDH-C gene was also observed in the populations of C. gachua only (Table-2).
Discussion
In subunit composition and expression of LDH-A or -B in a specific tissue, typical LDH zymograms of C. punctata, C. gachua and C. striatus are comparable to the reported electrophoretic profiles of several other teleosts [2,9,16]. Steady state patterns of C. punctata compared to its con-generic species, demonstrated the predominance of LDH-B4 in adult tissues such as brain, eye, heart, liver and kid-neys (Table-1). This may suggest that these tissues are comparatively aerobic, as reported previously [3,8,12]. However, the tissue restriction in LDH or its isoenzymes has been attributed to post translational modifications.
The presence of LDH-A4 or increase in LDH activity has been correlated with anaerobiosis or hypoxic conditions. Hence, the presence of this specific homotetramer or its subunits in various tissues of C. gachua and C. striatus may reveal the increase in rates of LDH activity due to either of the conditions described above which had been reported earlier in other fish species [8,17]. Therefore, the information on quantitative coding of these isoenzymes provided here may offer an explanation of the adaptive strategy adopted by the C. gachua and C. striatus under state of respiratory stress. While, higher levels of LDH-B in C. punctata seems to have favored least against the hypoxia/asphyxia induced alterations as an adaptive trait [13].
LDH zymograms as explained under results show the presence of two detectable activities in C. punctata and C. striatus that typically correspond to two loci of LDH namely LDH-A and LDH-B (Table-2). The inferred homotetrameric composition of the representative LDH activities of A and B loci and that of their random hetero-tetramers (-A1B3, -A2B2, -A3B1) is also confirmed by heat inactivation (results not shown here). Published evidence also reveals the existence of a third locus ‘C’ that is tem-porally expressed in fishes [18]. Previously it was referred to as E locus because it was known to predominantly function in regions of nervous system concerned with vision, but subsequently an orthologous expression was found in other tissue such as the liver and thus renamed as ‘C’ [1]. It is estimated that C-locus might have evolved as a result of duplication of B [2-3,8] whereas predecessor of A and B was a single gene of more remote origin.
Whitt and Maeda [19] have also reported that blind cave fish, Anoptichthys jordani, also lack the LDH-C gene functioning specially in the nervous system because of loss of visual structures and function in this fish. How-ever, other closely related species of characins and some other teleosts have been reported to lack function of this gene [19,20] that strongly suggests that LDH-C4 isoen-zyme is not an essential component for vision. Therefore, the lack of LDH-C gene in C. punctata and C. striatus may indicate that evolutionarily they are not of recent origin. They may be phylogenetically more close to the fishes belonging to super order Osteriophysi; and the spe-cies of Characiformes and Siluriformes that lack the func-tioning of LDH-C gene. However, unambiguous presence of LDH-C in the eye of C. gachua elucidates its evolu-tionary closeness with species of bony fishes belonging to class Acipenseriformes, Amiiformes, Anguilliformes, Myctophiformes, Perciformes, Pleuronectiformes, etc. where the existence of this gene has been correlated with the metabolism of photoreceptor cells in the retina [1,2].
Out of the three species of Channids selected here, C. gachua is a polyploid with 2n=78 [21]. As polyploidiza-tion is an important sympatric speciation process which marks an important evolutionary step- an autapomorphy for newly formed species [22]. Therefore, it may be pro-posed that during the course of evolution with the event like polyploidization or eventual synapomorphy, C. gachua might have evolved as a fish of generalized exis-tence against the selective or stochastic forces similar to few other groups of bony fishes. Reports are also available where due to differed evolutionary protocols, varia-tions in the level of gene expression have been demon-strated to contribute dissimilarity in individual tissue enzyme/ isoenzyme levels [12,16]; hence these variations can be utilized to understand the mode of evolution of regulatory genes controlling the steady state levels of these enzyme molecules and other related metabolic processes in the selected air-breathing teleosts [16]. Now, it appears that the significance of these characters (isoenzyme coding proposed here) in estimating the genetic di-vergence or evolution of different fish species at tissue level may be of great impact and further elaborative stud-ies should be designed in this direction to explore the sur-vivorship of air-breathing teleosts against the unknown selective forces acting during the course of evolution.
Acknowledgements
Thanks are due to the Chairman, Department of Zoology for providing the facilities and to the Department of Sci-ence and Technology (DST), Ministry of Science and Technolgy, New Delhi for granting the Research project under SERC-Fast Track Scheme for Young Scientists (FAST).
References
- Markert CL, Shaklee JB, Whitt GS. Evolution of a gene. Science 1975; 189: 102-114.
- Whitt GS. In: Isozymes: Current Topics in Biological and Medical Research. Rattazi, MC, Scandalios JG, Whitt GS eds. Vol. 10. Alan R Liss, New York 1983; pp. 1-40.
- Almeida-Val VMF, Luis Val A. Evolutionary trends of LDH isozymes in fishes. Comp Biochem Physiol 1993; 105B: 21-28.
- Coquelle N, Fioravanti E, Weik M, Vellieux F, Madern D. Activity, stability and structural studies of lactate dehydrogenases adapted to extreme thermal environ- ments. J Mol Biol 2007; 374(2): 547-562.
- Padhi BK, Khuda-Bukhsh AR. Lactate dehydrogenase isozyme patterns in developing stages of four species of Indian carps (Cyprinidae, Pisces). Natl Acad Sci Let- ters 1989; 12(2): 59-61.
- Rao MRK, Padhi BK, Khuda-Bukhsh AR. Lactate de- hydrogenase isozymes in fifty two species of teleostean fishes: Taxonomic significance of Ldh-C gene expres- sion. Biochem Syst Ecol 1989; 17(1): 69-76.
- Basu SK, Ganguly S, Sarkar SK, Basu TK. Ontogenetic and comparative analysis of LDH isozymes in the three species of Indian major carp. Ind J Exp Biol 1992; 30: 90-93.
- Masters CJ, Holmes RS. Hemoglobin, isoenzymes and tissue differentiation. American Elsevier publishing Co Inc, New York 1975.
- Champion MJ, Shaklee JB, Whitt GS. In: Isozymes, Developmental Biology. Markert CL ed. Vol. 3. Aca- demic Press, New York 1975; pp 417-439.
- Fisher SE, Shaklee JB, Ferris SD, Whitt GS. Evolution of five multilocus isozyme systems in the chordates. Genetica 1980; 52/53: 73-85.
- Avise JC. Systematic value of electrophoretic data. Syst Zool 1974; 23(4): 465-481.
- Buth DG, Murphy RW. The use of isozyme characters in systematic studies. Biochem Syst Ecol 1999; 27: 117-129.
- Ahmad R, Hasnain A. Ontogenetic changes and developmental adjustments in lactate dehydrogenase isozy- mes of an obligate air-breathing fish Channa punctatus during deprivation of air access. Comp Biochem Physiol 2005; 140 B: 271-278.
- Lowry OH, Rosenbrough HJ, Farr AL, Randall RS. Protein measurement with the folin phenol reagent. J Biol Chem 1951; 193: 265-275.
- Shaw CR, Prasad R. Starch gel electrophoresis of enzymes-a compilation of recipes. Biochem Genet 1970; 4: 279-320.
- Kettler MK, Ghent AW, Whitt GS. A comparison of phylogenies based on structural and tissue-expressional differences of enzymes in a family of teleost fishes (Salmoniformes: Umbridae). Mol Biol Evol 1986; 3: 485-498.
- Hochachka PW, McClelland GB, Burness GP, Staples JF, Suarez RK. Integrating metabolic pathway fluxes with gene-to-enzyme expression rates. Comp Biochem Physiol 1998; 120B: 17-26.
- Markert CL, Faulhaber I. Lactate dehydrogenase isozyme patterns of fish. J Exp Zool 1965; 159: 319-332.
- Whitt GS, Maeda FS. Lactate dehydrogenase gene function in the blind cave fish, Anoptichthys jordani, and other characins. Biochem Genet 1970; 4: 727-741.
- Nakano E, Whiteley AH. Differentiation of multiple molecular forms of four dehydrogenases in the teleost, Oryzias latipes, studies by disc electrophoresis. J Exp Zool 1965; 159: 167-180.
- Banerjee SK, Misra S, Banerjee S, Ray-Chaudhuri ST. Chromosomes numbers, genome sizes, cell volumes and evolution of snake-head fish (Family Channidae). J Fish Biol 1988; 33:781-789.
- Buth DG. The application of electrophoretic data in systematic studies. Ann Rev Ecol Syst 1984; 15: 501-522.