ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
Research Article - Biomedical Research (2018) Volume 29, Issue 12
Prediction system for heart disease using Naive Bayes and particle swarm optimization
Uma N Dulhare*
Muffakham Jah College of Engineering and Technology, Banjara Hills, Hyderabad, India
- *Corresponding Author:
- Uma N Dulhare
Muffakham Jah College of Engineering and Technology
Banjara Hills, Hyderabad, India
Accepted on May 21, 2018
DOI: 10.4066/biomedicalresearch.29-18-620
Visit for more related articles at Biomedical ResearchHeart attack disease is major cause of death anywhere in world. Data mining play an important role in health care industry to enable health systems to properly use the data and analytics to identify impotence that improves care with reduce costs. One of data mining technique as classification is a supervised learning used to accurately predict the target class for each case in the data. Heart disease classification involves identifying healthy and sick individuals. Linear classifier as a Naive Bayes (NB) is relatively stable with respect to small variation or changes in training data. Particle Swarm Optimization (PSO) is an efficient evolutionary computation technique which selects the most optimum features which contribute more to the result which reduces the computation time and increases the accuracy. Experimental result shows that the proposed model with PSO as feature selection increases the predictive accuracy of the Naive Bayes to classify heart disease.
Keywords
Heart disease, Naive based classifier, Particle swarm optimization, Feature selection.
Introduction
Angina, chest pain and heart attack are the symptoms of Coronary Heart Disease (CHD), Cardiovascular Diseases (CVDs) elucidated around one fourth of all deaths in India in 2008. In India, there could be 30 million CHD patients out of which 14 million are in urban and 16 million in rural areas. [1]. The heart attack increases due to smoking, lack of exercises, high blood pressure, high cholesterol, improper diet, high sugar levels etc. [2]. Early detection and treatment can keep heart disease from getting worse.
In the past few decades, medical data mining have played a important role to explore the hidden patterns which can be used for clinical diagnosis of any disease dataset [3].
Classification is one of the data mining technique to classify the patient class as normal or heart disease but classification use all attributes either relevant or irrelevant features which may reduce the classification performance. Feature subset selection is one of the dimensionality reduction techniques use to improve the accuracy. Particle swarm optimization is a feature selection technique which removes the redundant features to improve the classifier performance.
Our proposed model identifies the relevant features and removes the irrelevant features, to predict the heart disease effectively.
Related Work
Data mining plays an important role in the field of heart disease prediction. Medical Data mining has great potential like exploring the hidden patterns which can be utilized for clinical diagnosis of any disease dataset. Several data mining techniques are used in the diagnosis of heart disease such as Naive Bayes, Decision Tree, neural network, kernel density, bagging algorithm, and support vector machine showing different levels of accuracies. Naive Bayes is one of the successful classification techniques used in the diagnosis of heart disease patients.
Peter et al. [4] talked about a new feature selection method algorithm which is the hybrid method which combined CFS and Bayes theorem (CFS+Filter Subset Eval) and evaluated accuracy 85.5%.
Shouman [5] presented work by integrating k-means clustering with Naive Bayes using different initial centroid selection to improve the Naive Bayes accuracy for diagnosing heart disease patients and accuracy was 84.5%.
Rupali et al. [6] decision support in Heart Disease Prediction System (HDPS) is developed by using both Naive Bayesian Classification and Jelinek-Mercer smoothing technique. This Laplace smoothing is use to make an approximating function which attempts to capture important patterns in the data to avoid noise & accuracy is 86%.
Elma et al. [7] proposed a classifier with the distance-based algorithm K-nearest neighbour and statistical based Naive Bayes classifier (cNK) and achieved the accuracy 85.92% for heart disease dataset.
Background
This section provides the basic concepts of classifier as Naive Bayes and feature subset selection method as PSO.
Particle swarm optimization (PSO)
PSO is an Evolutionary Computation technique is proposed by Kennedy et al. in 1995 [8]. PSO is motivated by social behaviors such as bird flocking and fish schooling. In PSO population swarm consists of “n” particles, and the position of each particle stands for the potential solution in D-dimensional space. The particles change its condition based on three aspects: (1) To keep its inertia; (2) To change the condition according to its most optimist position; (3) To change the condition according to the swarm’s most optimist position.
In PSO [9], a population are encoded as particles in the search space dimensionality D.PSO starts with the random initialisation of a population of particles. Based on the best experience of one particle (pbest) and its neighbouring particles (gbest), PSO searches for the optimal solution by updating the velocity and the position of each particle; PSO is used as feature subset selection method due to its advantages:
• Simple and easy to implement.
• Continuous optimisation approach.
Naive Bayes classifier
Naive Bayes classifiers are a family of simple probabilistic classifiers based by using Bayes theorem with strong (Naive) independence assumptions between the features [10]. Naive Bayes classifiers are highly scalable by requiring a number of parameters linear for the number of features or predictors as variable in a learning problem. It is the simplest and the fastest probabilistic classifier especially for the training phase [11].
Feature selection
It is a process of removing the irrelevant and redundant features from dataset based on evaluation criterion which is use to improve accuracy. There are two approaches as individual evaluation and other one is subset evaluation.
The process of feature selection is classified into three broad classes. One is filter and another one is wrapper and third one is embedded method based on how the feature selection is deployed by supervised learning algorithm [3].
In this paper, we propose a model which uses Naive Bayes as classifier and PSO as Feature subset selection measure for prediction of heart disease.
Proposed system
In this section, we propose a methodology to improve the performance of Bayesian classifier for prediction of heart disease. Algorithm for our proposed model is shown below:
Algorithm 1: Heart disease prediction by using Bayes classifier and PSO.
Input: Heart disease dataset.
Output: Classify patient dataset into heart disease or not (normal).
Step 1: Read the dataset.
Step 2: Apply particle swarm optimization for feature selection.
Step 3: Remove the features with low value of PSO.
Step 4: Apply Naive Bayes classifier on relevant features.
Step 5: Evaluate the performance of NB+PSO model.
The above algorithm divided into two sections, section 1 (step 2 and step 3) performs processing and feature subset selection. In section 2 (step 4 and step 5) Naive Bayes is applied on relevant features data and evaluate the performance in terms of accuracy.
Accuracy=(No. of objects correctly classified/Total no. of objects in test set)
Cross validation technique used to split into training and testing data.
Experimental Results
In the proposed method, the accuracy of Naive Bayes classifier is improved by using the particle swarm optimization as feature selection. The dataset that is being used in the process contains the following 14 features including 1 class label and 270 instances with no missing values data collected from Statlog (heart) data set UCI repository which is balanced [12,13]. Here presence or absence of heart disease is predicted on the basis of age, sex, chest pain type, resting blood pressure, serum cholesterol, fasting blood sugar, resting electrocardiographic results, maximum heart rate achieved, exercise induced angina and old peak etc. which are given as follows (Table 1).
| Age | Age in years |
| Gender | Gender (male, female) |
| Cp | Chest pain type (typical angina, atypical angina, non-anginal pain, asymptomatic) |
| Trestbps | Resting blood pressure (in mmHg on admission to the hospital) |
| Chol | Serum cholesterol in mg/dl |
| Fbs | Fasting blood sugar>120 mg/dl (true, false) |
| Restecg | Resting electrocardiographic results (normal, abnormal, LVH) |
| Thalach | Maximum heart rate achieved |
| Exang | Exercise induced angina (yes, no) |
| Oldpeak | ST depression induced by exercise relative to rest |
| Slope | The slope of the peak exercise ST segment (unsloping, flat, downsloping) |
| Ca | Number of major vessels (0-3) colored by fluoroscopy |
| Thal | Normal, fixed defect, reversible |
| Num | Diagnosis of heart disease (yes, no) |
Table 1. 14 features including 1 class label and 270 instances.
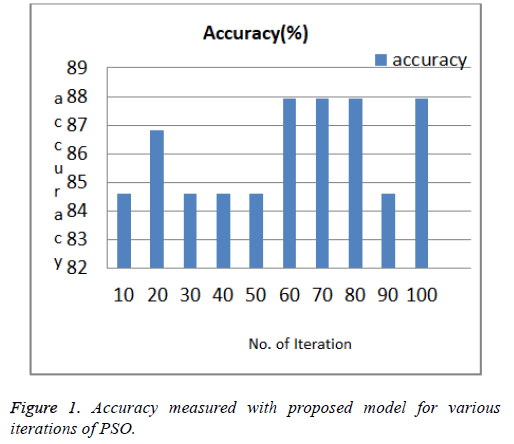
Open source statistical tool R with caret and PSO package used; for prediction. PSO searches out of 14, selects 7 featurescp, Restecg, Thalach, Exang, Old peak, Ca, Thal with 1 class. Result of proposed approach for various PSO search iteration as shown in Table 2 and Figure 1. In PSO optimization process, the result is obtained on convergence no. of iterations can be stopped if negligible deviation in convergence results. For example accuracy 87.91% recorded at max no. of iterations=100 as shown in Table 2.
| Max. no. of iterations | Selected feature subset | Accuracy (%) PSO+NB |
|---|---|---|
| 10 | 1, 2, 3, 4, 8, 9, 10, 13 | 84.62 |
| 20 | 2, 3, 4, 9, 10, 12, 13 | 86.81 |
| 30 | 2, 3, 4, 6, 9, 10, 12, 13 | 84.61 |
| 40 | 2, 3, 4, 6, 9, 10, 12, 13 | 84.61 |
| 50 | 3, 7, 9, 12 | 84.61 |
| 60 | 2, 3, 6, 8, 9, 12, 13 | 86.91 |
| 70 | 3, 7, 8, 9, 10, 12, 13 | 87.91 |
| 80 | 2, 6, 8, 9, 12, 13 | 87.01 |
| 90 | 2, 3, 4, 6, 8, 9, 10, 12, 13 | 84.62 |
| 100 | 3, 7, 8, 9, 10, 12, 13 | 87.91 |
Table 2. Various iterations of PSO for feature subset selection.
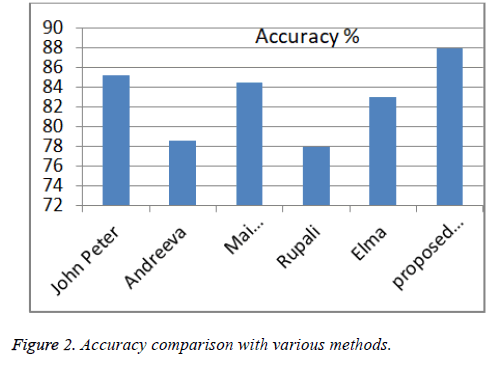
| S. no. | Author | Technique | Accuracy (%) |
|---|---|---|---|
| 1 | John Peter [4] | CFS and Bayes theorem | 85.5 |
| 2 | Polat [13] | AIS | 84.5 |
| 3 | Shouman [5] | Naive Bayes and K-means | 84.5 |
| 4 | Rupali [6] | Naive Bayes and Jelinek mercer by Laplace smoothing | 86 |
| 5 | Elma [7] | K-nearest neighbor and Naive Bayes | 82.96 |
| 6 | Our approach | PSO with NB | 87.91 |
Table 3. Accuracy comparison with various methods.
Comparison of other model with our proposed model as shown in Table 3 and Figure 2; our approach improves the accuracy which helps the doctor to advice the patient for further diagnosis process.
The predictive model with Naïve Bayes accuracy is 79.12% and result recorded by our proposed model Naive Bayes+PSO is 87.91%. Our proposed model improved the accuracy 8.79% as compared to existing system.
Proposed approach (NB+PSO) is compared with NB+GA. Using GA, accuracy is recorded as 86.29%, which is shown in Table 4 [14-18].
| Data set name | Approach | Accuracy (%) |
|---|---|---|
| Heart disease | Naive Bayes+GA | 86.29 |
| Naive Bayes+PSO | 87.91 |
Table 4. Accuracy comparison with GA and PSO.
Conclusion
It concluded that the proposed model is effective and efficient to improve the accuracy of the Naive Bayes classifier using the particle swarm optimization for feature subset selection which achieves similar or even better classification performance. The goal was successfully achieved by developing a novel algorithm maximizing the classification performance and minimizing the number of features. From simulation results, it analysed that this algorithm could automatically evolve a feature subset selection with a less number of features and increase classification performance than using all the features of a dataset. In the future, we want develop recommendation system for early prediction of heart disease diagnosis. Also the use of PSO for feature selection on datasets with a huge number of features can also be studied to note the various aspects of PSO in feature selection.
References
- Manoj B, Kumar G, Ramesh G, Sushil G. Emerging risk factors for cardiovascular diseases: Indian context. Indian J Endocrinol Metab 2013; 17: 806-814.
- Halaudi Daniel M. Prediction of heart disease using classification algorithms. WCSECS 2014; 22-24.
- Uma ND, Ayesha. Extraction of action rules for chronic kidney disease using Naive Bayes classifier. IEEE Int Conference Comput Intelligence Comput Res 2016.
- Jonh Peter T. Study and development of novel feature subsets selection framework for hard disease prediction. IJSRP 2012; 2.
- Mai S. Integrating Naive and clustering with a different initial centroid selection methods in the diagnosis of heart disease prediction. CS IT CSCP 2012; 125-137.
- Rupali RP MS. Heart disease prediction system using naive based and Jelmeck Mercer smoothing. IJARCCE 2014; 3: 6787-6792.
- Elama Zannatul F. Combination of Naive Bayes classifier and K-NN in the classification based classification models. Computer Inform Sci 2013; 6: 48-56.
- Kennedy J, Eberhart R. Particle swarm optimization. In Proc IEEE Int Conf Neural Netw 1995; 4: 1942-1948.
- Bing X, Mengie Z, Will NB. Particle swarm optimization in future selection in classification: a multi objective approach. In IEEE Transaction on Cybernetics 2013.
- Wikipedia. Naïve Bayes classifier. Wikipedia 2018.
- Fayeza A, Uma ND. A machine learning based approach for opinion mining on social network data. LNNS Springer Proceedings of IC3T 2016; 5: 135-148.
- Machine Learning Repository. 436 Data sets. UCI 1989.
- Polat K, Sahan S, Kodaz H, Günes S. A new classification method to diagnose heart disease: Supervised artificial immune system. In Proceedings of the Turkish Symposium on Artificial Intelligence and Neural Networks (TAINN) 2005; 2186-2193.
- Shi Y, Eberhart R. A modified particle swarm optimizer. In Proc IEEE Int CEC 1998; 69-73.
- Bing X, Mengie Z, Will NB. Particle swarm optimization in feature selection as classification novel initiation and updating mechanism. In Appl Soft Comput 2014; 261-276.
- Guyon AE. An introduction to variable and feature selection. J Machine Learning Res 2003; 1157-1182.
- Zang H. The optimality of Naïve Bayes. Am Assoc Artificial Intelligence 2004.
- Aha D, Kibler. Instance bays prediction of heart disease presence with the Cleveland data base. Technical Rep ICS-TR 1988.