ISSN: 0970-938X (Print) | 0976-1683 (Electronic)
Biomedical Research
An International Journal of Medical Sciences
- Biomedical Research (2015) Volume 26, Issue 1
NAD(P)H: Quinone Oxidoreductase 1 inducer activity of some enaminone derivatives.
1Department of Pharmacognosy, College of Pharmacy, King Saud University, P.O. Box 2457, Riyadh 11451, Kingdom of Saudi Arabia
2Jacqui Wood Cancer Centre, Division of Cancer Research, Medical Research Institute, University of Dundee, Dundee DD1 9SY, United Kingdom
3Departments of Medicine and Pharmacology and Molecular Sciences, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA
- *Corresponding Author:
- M.M. Ghorab
Section of Applied Organic Chemistry
Department of Pharmacognosy
College of Pharmacy, King Saud University
P.O. Box 2457, Riyadh 11451
Kingdom of Saudi Arabia.
Accepted July 07 2014
The present work reports the synthesis of some enaminone derivatives bearing the biologically active 3,4-dimethoxyphenyl (3) or 3,4,5-trimethoxyphenyl moieties (5 and 7), respectively. The trimethoxybenzene moiety has been previously reported to confer cytotoxic activity. However, we found that, at high micromolar concentrations, the new compounds have the ability to weak-ly induce the cytoprotective enzyme NQO1. This is most likely due to their electrophilic cyclo-hexenone functionality, a well-established structural feature of NQO1 inducers. The structure of the newly synthesized compounds was confirmed on the basis of elemental analyses, IR, 1H-NMR, 13C-NMR spectra.
Keywords
Synthesis, enaminones, NQO1, electrophilicity, cytoprotection
Introduction
Enaminones are a group of organic compounds carrying the conjugated system N- C= C- C= O. The literature has reported information about the chemistry of enaminones, their physicochemical properties and biological activities [1-4]. In addition, cyclohex-2-enone has a wide range of biological activities such as anticancer [5] and antimicrobi-al activities [6]. On the other hand, enaminones have been extensively used as key intermediates in organic synthesis [7-12] and the chemistry of these compounds has been re-viewed [13]. In particular they have been employed as synthons of a wide variety of biologically active heterocy-clic compounds [14], as pharmaceutical agents with anti-cancer [15], antibacterial [16], anti-inflammatory [17] and other therapeutic activities [18-20]. During the past decade we have been involved in a program aimed at exploring the potential of enaminone as building blocks for heteroaro-matics [21], and have successfully synthesized quinoline derivatives7-12 utilizing enaminones as starting materials. Based on the above information and as a continuation of previous work on anticancer agents [22-27], we report the synthesis of some new enaminone derivatives.
Experimental
The starting materials cyclohexane-1,3- dione, 5,5- dime-thylcyclohexane-1,3- dione, 3,4- dimethoxyaniline, and 3,4,5- trimethoxyaniline were purchased from Sigma- Aldrich. Melting points were determined on an electro-thermal melting point apparatus (Stuart Scientific, Stone), and were uncorrected. Precoated Silica gel plates (Kiesel gel 0.25 mm, 60 G F 254, Merck) were used for thin layer chromatography (TLC). The developing solvent system was chloroform / methanol (10 : 3) and the spots were detected by ultraviolet light. Infrared (IR) spectra (KBr disc) were recorded on FT- IR spectrophotometer (Perkin Elmer) at the Research Center, College of Pharmacy, King Saud University, Saudi Arabia. 1H-NMR spectra were scanned in dimethylsulfoxide (DMSO-d6) on a NMR spectrophotometer (Bruker AXS Inc.) operating at 500 MHz for 1H and 125.76 MHz for 13C at the aforemen-tioned Research Center. Chemical shifts are expressed in d- values (ppm) relative to tetramethylsilane (TMS) as an internal standard. Exchangeable protons were confirmed by addition of a drop of D2O. Elemental analyses were done on a model 2400 CHNSO analyzer (Perkin Elmer).
Results
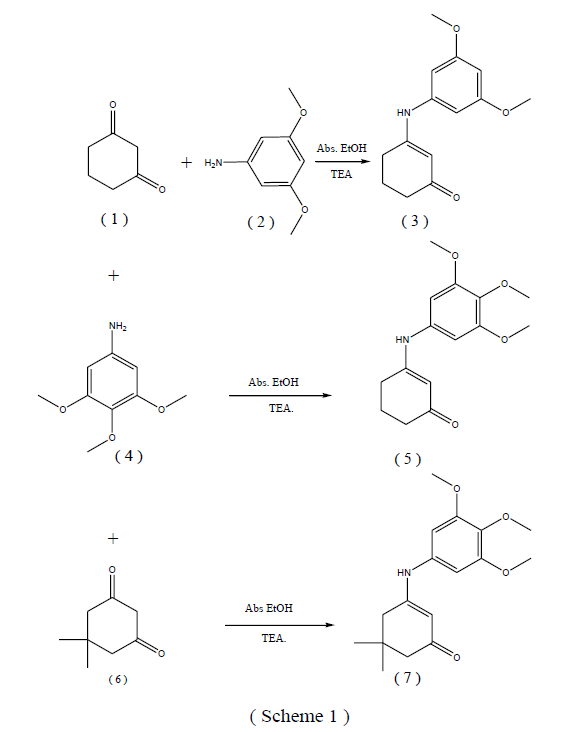
Synthesis of 3-(3,5-dimethoxyphenylamino) cyclohex-2- enone (3).
A mixture of cyclohexane -1,3- dione 1 (1.22g, 0.01 mole) and 3,5- dimethoxyaniline 2 (1.53g, 0.01 mole) in absolute ethanol (30 mL) containing 3 drops of triethylamine was refluxed for 8h. The reaction mixture was cooled and the solid obtained was recrystallized from dioxane to give 3. Yield % 94; m.p. 138-140 ºC; IR (KBr, Cm-1): 3298 (NH), 3086 (CH arom.), 2972, 2876 (CH aliph.), 1652 (C=O). 1H-NMR spectrum in (DMSO-d6): 1.39,1.87, .99 [m, 6H, 3CH2 cyclo.], 3.70 [s, 6H, 2 OCH3], 5.7 [s, 1H, CH cyclo.], 5.9 – 6.3 [m, 3H, Ar-H], 9.7 [ s, 1H, NH, D2Oexchangeable]. 13C-NMR spectrum in (DMSO-d6): 19.6, 28.3, 38.8, 56.4 (2), 91.2, 92.6, 101.6, 145.9, 161.4, 163.7 (2), 200.6 (C=O). Anal. Calcd for C14H17NO3 (247.29): C, 68.00; H, 6.93; N, 5.66. Found: C, 68.31; H, 6.64; N, 5.93.
Hepa1c1c7 cells (104 per well) were grown in 96-well plates for 24 h. After that, the cell culture medium was replaced with fresh medium containing serial dilutions of enaminones. Cells were grown for a further 48h, and lysed with digitonin. The specific activity of NQO1 was determined in cell lysates using menadione as a substrate. Mean values for 8 replicate wells are shown for each data point. The standard deviation in each case was <5% of the value.
Synthesis of 3-(3,4,5-trimethoxyphenylamino) cyclohex- 2-enone ( 5 ).
A mixture of cyclohexane-1,3-dione 1 (1.12g, 0.01 mole) and 3,4,5- trimethoxyaniline 4 (1.83g, 0.01 mole) in absolute ethanol ( 30 mL) containing 3 drops of triethylamine was refluxed for 6h. The reaction mixture was cooled and the solid obtained was recrystallized from ethanol to give 5. Yield % 89; m.p. 205-207 ºC; IR (KBr, Cm-1): 3270 (NH), 3100 (CH arom.), 2966, 2836 (CH aliph.), 1653 (C=O). 1H-NMR spectrum in (DMSO-d6): 1.8- 2.4 [m, 6H, 3CH2 cyclo.], 3.70, 3.78 [2s, 9H, 3 OCH3], 5.2 [s, 1H, CH cyclo.], 6.4 [s, 2H, Ar-H], 8.7 [s, 1H, NH, D2O- exchangeable]. 13C-NMR spectrum of 5 in (DMSO-d6): 21.5, 28.4, 36.3, 55.8 (2), 60.0, 98.0 (2), 101.1, 134.4, 135.6, 153.0 (2), 162.2, 195.7 (C=O). Anal. Calcd for C15H19NO4 (277.32): C, 64.97; H, 6.91; N, 5.05. Found: C, 64.69; H, 6.59; N, 5.31, melting point, IR, 1HNMR, 13C-NMR and elemental analysis as reported by us7.
Biological evaluation
The NQO1 inducer activity was determined using a quantitative microtiter plate assay [28]. Hepa1c1c7 cells were grown in αMEM supplemented with 10% (v/v) fetal bovine serum that had been heat- and charcoal-inactivated. Cells were routinely maintained in a humidified atmosphere at 37 °C, 5% CO2. For each experiment, cells (104 per well) were plated in 96-well plates. After 24 h, the cell culture medium was replaced with fresh medium containing enaminones, and the cells were grown for a further 48 h. Eight replicates of 8 serial dilutions of each compound were used. Compounds were prepared as stock solutions in DMSO, and then diluted in the cell culture medium 1:1000. The final concentration of DMSO in the medium was maintained at 0.1% (v/v). At the end of the 48 h exposure period, cells were lysed for 30 min at 25 °C in digitonin (0.8 g/L, pH 7.8). The specific activity of NQO1 was evaluated in cell lysates using menadione as a substrate. Protein concentrations were determined in each well by the BCA protein assay (Thermo Scientific). Sulforaphane, a classical NQO1 inducer, served as a positive control.
Discussion
The aim of the present work was the design, synthesis and structure elucidation of some enaminone derivatives carrying a biologically active 3,5-dimethoxyphenyl moiety 3 and 3,4,5-trimethoxyphenyl moieties 5 and 7 with expected anticancer activity (Scheme 1). 3-(3,5- Dimethoxyphenyl-amino)cyclohex-2-enone 3 was obtained in good yield via reaction of cyclohexane-1,3-dione 1 with 3,5- dimethoxyaniline 2 in refluxing ethanol containing a few drops of triethylamine as a catalyst (Scheme 1). The structure of compound 3 was supported by elemental analysis, IR, 1H-NMR, 13C-NMR spectra and xray data. IR spectrum of 3 revealed the presence of characteristic bands for NH at 3310 Cm-1, (CH arom.) at 3096 Cm-1, (CH aliph.) at 2977, 2866 Cm-1 and (C=O ) at 1647 Cm-1. Also, 1H-NMR spectrum in (DMSO-d6) indicated the presence of a signals at 3.71 ppm which could be assigned to two methoxyl groups, 5.7 ppm due to CH cyclo., and 9.7 ppm for NH of enaminone 3. 13CNMR spectrum of 3 in (DMSO-d6) showed signals at 19.6, 28.3, 38.8, 56.4 (2), 91.2, 92.6 (2), 101.6, 145.9, 161.4, 163.7 (2), 200.6 (C=O).
In addition, interaction of 1 with 3,4,5-trimethoxyaniline 4 in refluxing ethanol afforded the corresponding 3- (3,4,5-trimethoxyphenylamino)-cyclohex-2-enone 5 in good yield. The structure of 5 was confirmed from its microanalysis, IR, 1H-NMR, 13C-NMR and x-ray data. Thus, IR spectrum of 5 exhibited the presence of charac-teristic bands for NH, CH aromatic, CH aliphatic, and (C=O). 1H-NMR spectrum of 5 revealed signals at 3.70, 3.78 ppm corresponding to three methoxy groups, 5.2 ppm due to CH cyclo and 8.7 ppm for NH group. 13C-NMR spectrum of 5 in (DMSO-d6) showed a signal at 195.7 (C=O). On the other hand, condensation of 5,5- dimethyl-cyclohexane-1,3-dione 6 with 3,4,5- trimethox-yaniline 4 yielded the corresponding 3-( 3,4,5- trimethox-yphenylamino)-5,5-dimethylcyclohex-2-enone 7 in good yield [23]. The structure of compound 7 was confirmed on the basis of elemental analysis, IR, 1H-NMR, 13C-NMR, and x-ray analysis. The IR spectrum of 7 showed bands for (NH), (CH aromatic), (CH aliphatic), and (C=O). Also, the 1H-NMR spectrum in (DMSO-d6) indi-cated the presence of a singlet at 8.7 ppm which could be assigned to NH of enaminone 3.
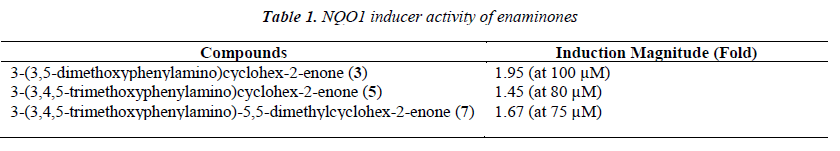
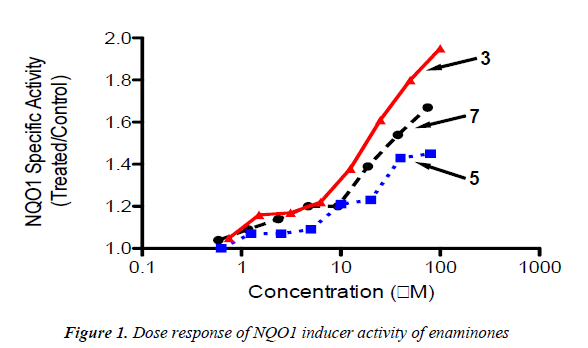
Based on the electrophilicity of the cyclohexenone func-tionality, we tested the possibility that enaminones may be able to induce the cytoprotective enzyme NAD(P)H:quinone oxidoreductase 1 (NQO1). Indeed, the cyclopentenone prostaglandins are well-known endoge-nous activators of nuclear factor-erythroid 2 p45-related factor 2 (Nrf2), the main transcription factor which regu-lates the basal and inducible expression of NQO1 [29-32]. Using a quantitative bioassay in murine Hepa1c1c7 cells [28,33], we found that all three enaminones are weak in-ducers of NQO1 (Table 1 and Figure 1). Exposure of the cells to the enaminones for 48 h led to a dose-dependent upregulation of the specific activity of NQO1. The magni-tude of induction among the three compounds was simi-lar, with compound 3 being the most potent, and com-pound 5 being the least potent inducer. No cytotoxicity was observed at any of the tested concentrations of the enaminones.
The 3,4,5-trimethoxyphenyl moiety is an important struc-tural feature for binding to tubulin and inhibition of mi-crotubule polymerization, and is also present in the struc-ture of colchicine [34]. In contrast, the similarity in in-ducer potency among the three enaminones indicates that the 3,4,5-trimethoxy substitution does not play a signifi-cant role for NQO1 induction as compound 3 lacks one of the methoxyl groups. This result points out to the im-portance of the cyclohexenone functionality, a feature shared by all three compounds. The electrophilic enone functionality was early recognized as a critical feature within a structurally diverse array of inducers of NQO1, due to the ability of enones to form Michael adducts with sulfhydryl groups [35]. By analogy with other NQO1 in-ducers bearing enone groups, such as phenylpropenoids, chalcones, curcuminoids and coumarins [36-38], we pro-pose that the enaminones activate transcription factor Nrf2 by reacting with cysteine sensors of its major nega-tive regulator, the ubiquitin ligase substrate adaptor Kelch-like ECH-associated protein 1 (Keap1). Under ho-meostatic conditions, Keap1 continuously targets Nrf2 for ubiquitination and proteasomal degradation [39-42]. Cys-teine modification of Keap1 leads to a loss of its repressor function, ultimately resulting in Nrf2 stabilization, nucle-ar translocation, and activation of its target genes, such as NQO1.
Acknowledgements
The authors would like to extend their sincere apprecia-tion to the Deanship of Scientific Research at King Saud University for funding of this research through the Re-search Group Project no. RGP-VPP-262. We are also grateful to Cancer Research UK (C20953/A10270) for financial support.
References
- Cox DS, Gao H, Raje S, Scott KR, Eddington ND, (2001) Enhancing the permeation of marker com- pounds and enaminone anticonvulsants across Caco-2 monolayers by modulating tight junctions using zonula occludens toxin. Eur. J. Pharm. Biopharm. 52, 45–150.
- Eddington ND, Cox DS, Khurana M, Salama NN, Sta- bles JP, Harrison SJ, Negussie A, Taylor RS, Tran UQ, Moore JA, et al. (2003) Synthesis and anticonvulsant activity of enaminones Part 7. Synthesis and anticon- vulsant evaluation of ethyl 4-[(substituted phenyl) ami- no]-6-methyl-2-oxocyclohex-3-ene-1-carboxylates and their corresponding 5-methylcyclohex-2-enone deriva- tives. Eur. J. Med. Chem. 38, 49–64.
- Edafiogho IO, Ananthalakshmi KV, Kombian SB, (2006) Anticonvulsant evaluation and mechanism of action of benzylamino enaminones. Bioorgan. Med. Chem. 14, 5266–5272.
- Ananthalakshmi KV, Edafiogho IO, Kombian SB, (2006) Concentration-dependent effects of anticonvul-sant enaminone methyl 4-(4'-bromophenyl) aminocy- clohex-3-en-6-methyl-2-oxo-1-oate on neuronal excita- bility in vitro. Neuroscience. 141, 345-56.
- Claire LA, Gareth AM, Michela P, Robin GP, Ian JS, Tanja T, Roger CW, Katharine FW, Natasha SW, (2010) The synthesis of 2- oxyalkyl- cyclohex-2- enones, related to the bioactive natural products COTC and anthraminone A, which possess anti-tumour prop- erties, Tetrahedron, 66, 9049-9060.
- Kandhavelu M, Paturu L, Mizar A, Mahmudov K, Kopylovich M, Karp M, Yli-Harja O, Pombeiro A, Ri- beiro A, (2012) Synthesis, characterization and antimi- crobial activity of arylhydrazones of methylene active compounds, Pharmaceutical Chemistry Journal; 46, 157.
- Alsaid SM, , Mahmoud SB, Ghorab MM, (2011) Novel quinolines bearing a biologically active trimethoxy- phenyl moiety as a new class of antitumor agents, Arzneimittelfoorschung, 61, 527- 53.
- Saleh I, Alqasoumi AM, Al-Taweel AM, Alafeefy MM, Hamed EN, Ghorab MM, (2009) Synthesis and biological evaluation of 2-amino-7,7-dimethyl 4- substituted-5-oxo-1-(3,4,5-trimethoxy)-1,4,5,6,7,8- hexahydro-quinoline-3-carbonitrile derivatives as po- tential cytotoxic agents, Bioorg. Med. Chem. Lett. 19, 6939- 6942.
- Ghorab MM, Ragab FA, Noaman E, Heiba HI, El- Hossary EM, (2008) Utility of 4-(5,5-dimethyl-3-oxo- cyclohex-1-enylamino)benzenesulfonamide in the syn- thesis of novel quinolines as possible anticancer and radioprotective agents. Arzneimittelforschung. 58, 35-41.
- Ghorab MM, Ragab FA, Noaman E, Heiba HI, El- Hossary EM. S (2007) ynthesis of some novel quino- lines and pyrimido [4,5-b] quinolines bearing A sulfon- amide moiety as potential anticancer and radioprotec- tive agents. Arzneimittelforschung. 57, 795-803.
- Ghorab MM, Ragab FA, Hamed MM. Design, (2009) synthesis and anticancer evaluation of novel tetrahy- droquinoline derivatives containing sulfonamide moie- ty. Eur J Med Chem. 44, 4211-7.
- Alqasoumi SI, Al-Taweel AM, Alafeefy AM, Noaman E, Ghorab MM, (2010) Novel quinolines and pyrim- ido[4,5-b]quinolines bearing biologically active sulfon- amide moiety as a new class of antitumor agents, Eur. J. Med. Chem. 45, 738- 744.
- Ghorab MM, Alsaid MS, El-Hossary EM, (2011) In vitro cytotoxic evaluation of some new heterocyclicsulfonamide derivatives, J. Heterocyclic. Chem, 48, 563-571.
- Ghorab MM, Ragab FA, Hamed MM, (2010a) Synthe-sis and docking studies of some novel quinoline deriva- tives bearing a sulfonamide moiety as possible anti- cancer agents, Arzneimittelforschung, 60, 141-148.
- Kouznetsov VV, Vladimir A, Rojas R, Fernando Y, Vargas M, Leonor P, Gupta M, (2012) Simple C-2- Substituted Quinolines and their anticancer activity, lLetters in Drug Design & Discovery, 9, 680-686.
- Eswaran S, Adhikari AV, Chowdhury IH, Pal NK, Thomas KD, (2010) New quinoline derivatives: syn- thesis and investigation of antibacterial and antituber- culosis properties, Eur J Med Chem.; 45, 3374-83.
- Chia EW, Pearce AN, Berridge MV, Larsen L, Perry NB, Sansom CE, Godfrey CA, Hanton LR, Lu GL, Wal-ton M, Denny WA, Webb VL, Copp BR, Harper JL,
(2008) Synthesis and anti-inflammatory structure- activity relationships of thiazine-quinoline-quinones: in- hibitors of the neutrophil respiratory burst in a model of acute gouty arthritis., Bioorg Med Chem. 16, 9432-42. - Ismail MM, Ghorab MM, Noaman E, Ammar YA,Heiba HI, Sayed MY (2007) Novel synthesis of pyrrolo [2,3-d] pyrimidines bearing sulfonamide moieties as potential antitumor and radioprotective agents. Arzneimittelforschung 56, 301-8.
- Ghorab MM, Ragab FA, Alqasoumi SI, Alafeefy AM, Aboulmagd SA, (2010b) Synthesis of some new pyra- zolo[3,4-d]pyrimidine derivatives of expected anti- cancer and radioprotective activity. Eur J Med Chem. 45, 171-8.
- Sankaran M, Kumarasamy C, Chokkalingam U, Mohan PS, (2010) Synthesis, antioxidant and toxicological study of novel pyrimido quinoline derivatives from 4- hydroxy-3-acyl quinolin-2-one., Bioorg Med Chem Lett.; 20, 7147-51.
- Abdel- Gawad SM, El- Gaby MSA, Heiba HI, Aly HM, Ghorab MM, (2205) Synthesis and radiation sta- bility of some new biologically active hydroquinoline and pyrimido [4,5-b] quinoline derivatives, J. Chin. Chem. Soc, 52, 1227-1236.
- Ghorab MM, AlSaid MS, (2012a) Anticancer activity of novel indenopyridine derivatives,Arch. Pharm. Res, 35, 987-994.
- AlSaid MS, Ghorab MM, Nissan YM, (2012) Dapson in heterocyclic chemistry, part VIII: synthesis, molecu- lar docking and anticancer activity of some novel sul- fonylbiscompounds carrying biologically active 1,3- dihydropyridine, chromene and chromenopyridine moieties, Chemistry Central Journal, 6, 64.
- Ghorab MM, AlSaid MS, (2012b) Antitumor activity of novel pyridine, thiophene and thiazole deriva- tives,Arch. Pharm. Res, 35, 965-973.
- Ghorab MM, Al-Said MS, Nissan YM, (2012c) Dapson in heterocyclic chemistry, part V: synthesis, molecular docking and anticancer activity of some novel sul- fonylbiscompounds carrying biologically active dihy- dropyridine, dihydroisoquinoline, 1,3-dithiolan, 1,3- dithian, acrylamide, pyrazole, pyrazolopyrimidine and benzochromenemoieties., Chem Pharm Bull (Tokyo). 60, 1019-28.
- Ghorab MM, AlSaid MS, Nissan YM, (2012d) Dapson in heterocyclic chemistry Part VI: Synthesis and mo- lecular docking of some novel sulfonebiscompounds of expected anticancer activity, Arzneimittelforschung, 62, 497-507.
- Emad MK, El-Sawy ER, Howaida IA, (2012) Synthe- sis, antimicrobial, and antiviral activities of some new 5-sulphonamido-8-hydroxyquinoline derivatives, Ar- chiv Pharma Res., 35, 955-964.
- Prochaska HJ, Santamaria AB. (1988) Direct measure- ment of NAD(P)H:quinone reductase from cells cultured in microtiter wells: a screening assay for anticarcinogen- ic enzyme inducers. Anal Biochem. 169, 328-36.
- Levonen AL, Landar A, Ramachandran A, Ceaser EK, Dickinson DA, Zanoni G, Morrow JD, Darley-Usmar VM. (2004) Cellular mechanisms of redox cell signal- ling: role of cysteine modification in controlling antiox- idant defenses in response to electrophilic lipid oxida- tion products. Biochem J, 378 (Pt 2): 373-82.
- Zhang X, Lu L, Dixon C, Wilmer W, Song H, Chen X, Rovin BH. (2004) Stress protein activation by the cy- clopentenone prostaglandin 15-deoxy-delta12, 14- prostaglandin J2 in human mesangial cells. Kidney Int. 65, 798-810.
- Kim JW, Li MH, Jang JH, Na HK, Song NY, Lee C, Johnson JA, Surh YJ. (2008) 15-Deoxy-Delta (12,14)- prostaglandin J(2) rescues PC12 cells from H2O2- induced apoptosis through Nrf2-mediated upregulation of heme oxygenase-1: potential roles of Akt and ERK1/2. Biochem Pharmacol, 76, 1577-89.
- Tsujita T, Li L, Nakajima H, Iwamoto N, Nakajima- Takagi Y, Ohashi K, Kawakami K, Kumagai Y, Free- man BA, Yamamoto M, Kobayashi M. (2011) Nitro- fatty acids and cyclopentenone prostaglandins share strategies to activate the Keap1-Nrf2 system: a study using green fluorescent protein transgenic zebrafish. Genes Cells. 16, 46-57.
- Fahey JW, Dinkova-Kostova AT, Stephenson KK, Tal- alay P. (2004) The "Prochaska" microtiter plate bioas- say for inducers of NQO1. Methods Enzymol. 382, 243-58.
- Assadieskandar A, Amini M, Ostad SN, Riazi GH, Cheraghi-Shavi T, Shafiei B, Shafiee A. (2013) Design, synthesis, cytotoxic evaluation and tubulin inhibitory activity of 4-aryl-5-(3,4,5-trimethoxyphenyl)-2- alkylthio-1H-imidazole derivatives. Bioorg Med Chem. 21, 2703-9.
- Talalay P, De Long MJ, Prochaska HJ. (1988) Identifi- cation of a common chemical signal regulating the in- duction of enzymes that protect against chemical car- cinogenesis. Proc Natl Acad Sci U S A. 85, 8261-5.
- Dinkova-Kostova AT, Abeygunawardana C, Talalay P, (1998) Chemoprotective properties of phenylprope- noids, bis(benzylidene)cycloalkanones, and related Mi- chael reaction acceptors: correlation of potencies as phase 2 enzyme inducers and radical scavengers. J Med Chem. 41, 5287-96.
- Dinkova-Kostova AT, Talalay P, (1999) Relation of structure of curcumin analogs to their potencies as in- ducers of Phase 2 detoxification enzymes. arcinogen- esis 20, 911-4.
- Dinkova-Kostova AT, Massiah MA, Bozak RE, Hicks RJ, Talalay P, (2001) Potency of Michael reaction ac- ceptors as inducers of enzymes that protect against car- cinogenesis depends on their reactivity with sulfhydryl groups. Proc Natl Acad Sci U S A. 98, 3404-9.
- Hayes JD, McMahon M, Chowdhry S, Dinkova- Kostova AT, (2010) Cancer chemoprevention mecha- nisms mediated through the Keap1-Nrf2 pathway. An- tioxid Redox Signal. 13, 1713-48.
- Villeneuve NF, Lau A, Zhang DD. (2010) Regulation of the Nrf2-Keap1 antioxidant response by the ubiqui-tin proteasome system: an insight into cullin-ring ubiq- uitin ligases. Antioxid Redox Signal. 13, 1699-712.
- Baird L, Dinkova-Kostova AT. (2011) The cytoprotec- tive role of the Keap1-Nrf2 pathway. Arch Toxicol. 85, 241-72.
- Suzuki T, Motohashi H, Yamamoto M. (2013) Toward clinical application of the Keap1-Nrf2 pathway. Trends Pharmacol Sci. 34, 340-6.